Back to Journals » OncoTargets and Therapy » Volume 13
Prognostic Nomogram for Patients with Radical Surgery for Non-Metastatic Colorectal Cancer Incorporating Hematological Biomarkers and Clinical Characteristics
Authors Long P , Zang Y, Wang H , Liang X, Xie X , Han Z , Lin D , Wang Z, Huang S, Chen C
Received 2 December 2019
Accepted for publication 17 February 2020
Published 9 March 2020 Volume 2020:13 Pages 2093—2102
DOI https://doi.org/10.2147/OTT.S240843
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Peiyun Long,1,* Youya Zang,1,* Huan Wang,1 Xiumei Liang,1 Xuekun Xie,1 Zhiwei Han,1 Dongyi Lin,1 Zongyu Wang,1 Shan Huang,2 Chuang Chen1
1Department of Oncology, Guangxi Medical University Cancer Hospital, Nanning, Guangxi, People’s Republic of China; 2Department of Oncological Surgery, Guangxi Medical University Cancer Hospital, Nanning, Guangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chuang Chen
Guangxi Medical University Cancer Hospital, No. 71 Hedi Road, Qingxiu District, Nanning, Guangxi 530021, People’s Republic of China
Tel/Fax +86 771 533 0622
Email [email protected]
Background: There is a large difference in postoperative survival in patients with non-metastatic colorectal cancer. We aimed to develop nomograms incorporating both hematological biomarkers and clinical characteristics to predict overall survival (OS) in patients with radical surgery for non-metastatic colorectal cancer.
Methods: A retrospective analysis was performed on date from 508 patients who underwent radical resection of colorectal cancer at the Affiliated Tumor Hospital of Guangxi Medical University from December 2011 to December 2015. Simple random sampling was performed by dividing these patients into a training set (n=355) and validation set(n=153), which yielded a 7:3 ratio in the sample sizes between these groups. Based on COX regression analysis of the results from the training cohort, a nomogram was developed to predict the three-year and five-year overall survival rate, and internal verification was also performed. The nomogram prediction accuracy and discriminating ability were evaluated by Harrell’s C-index (C-index), calibration curves and were compared with the colorectal cancer TNM staging system.
Results: We found that age, degree of differentiation, T stage, N stage, neurological invasion, neutrophils, monocytes, HGB, and LDH were independent risk factors for predicting OS in patients with colorectal cancer. In the training cohort, the C index was 0.796 (95% CI: 0.761– 0.831). In the validation cohort, the C index was 0.671 (95% CI: 0.656– 0.686).The nomogram showed a stronger predictive ability than did TNM staging. Decision curve analysis showed that the nomogram had value in terms of clinical application.
Conclusion: Our nomogram combined hematological biomarkers and clinical characteristics and was highly effective in predicting OS in patients with non-metastatic colorectal cancer. Hence, our nomogram may provide a reference tool for clinicians to guide individualized treatment and follow-ups for patients with colorectal cancer.
Keywords: colorectal cancer, prognostic, mortality, nomogram
Introduction
Colorectal cancer (CRC) is the third most common malignant tumor in the world and the second leading cause of cancer-related death worldwide.1 In China, there were an estimated 376,300 newly diagnosed CRC cases and 191,000 CRC-related deaths in 2015.2 Compared with rates in developed countries such as the United States and Japan, the incidence and mortality of colorectal cancer in China have increased compared to those in previous years.3 Despite considerable progress in surgery and chemotherapy in recent years, the prognosis of CRC remains poor. Currently, the American Joint Committee on Cancer (AJCC) TNM staging system is a clinically and widely used staging system, This system has value in predicting prognosis and in guiding treatment for patients with CRC.4 However even if patients are in the same stage and if similar treatment strategies are used, survival outcomes may vary widely. Given the limitations of the TNM staging system, establishing a better model to predict the prognosis of patients undergoing radical resection of CRC is particularly important.
Obviously, due to the biological heterogeneity of tumors, there are many factors affecting the prognosis of colorectal cancer.5 In recent years, research has shown that blood biomarkers are associated with prognosis of colorectal cancer. These biomarkers include neutrophils, monocytes, lymphocytes, hemoglobin (HGB), lactate dehydrogenase (LDH), and other hematological biomarkers.6,7 However, few studies have conducted a comprehensive discussion of these hematological biomarkers. Hence, the value of these markers in determining the prognosis or the survival rate after CRC surgery should be further evaluated. Combining the prognostic factors of these hematological biomarkers, makes it possible to further improve the accuracy of CRC model prediction.
A nomogram is a reliable statistical model that establishes a graphical predictive tool to predict tumor prognosis by incorporating and screening for risk factors for tumorigenesis. A number of studies have shown that nomograms are prognostic in a variety of cancer populations and are more accurate than traditional TNM staging systems. These populations include patients with CRC, liver cancer, gastric cancer, and nasopharyngeal carcinoma.8–11 Given the individualized predictive power of this statistical tool. our present study combined hematological biomarkers and clinical features to establish and validate a nomogram for predicting overall survival (OS) in patients undergoing CRC radical surgery, providing a reference tool for clinicians to guide individualized treatment and follow-ups for CRC patients.
Materials and Methods
Study Population
This study retrospectively analyzed 508 patients with radical resection of colorectal cancer at the Guangxi Medical University Cancer Hospital from December 2011 to December 2015. Simple random sampling was performed by dividing these patients into a training cohort (n=355) and validation cohort (n=153), yielding a 7:3 ratio between these two groups. The inclusion criteria were as follows: (1) Patients were older than 18 years; (2) patients underwent radical resection(radical surgery is defined as complete tumor resection and pathological examination confirming R0 resection)without intestinal obstruction, perforation, hemorrhage, or other complications. They had histopathologically confirmed colorectal cancer, including adenocarcinoma, showing mucus and cellular cancer; and (3) laboratory tests were completed before surgery. The exclusion criteria were as follows: (1) Patients who received anti–inflammatory drugs (including antibiotics) or immunosuppressive therapy (including steroids) within there months before surgery, or who had chronic inflammatory diseases including infection and autoimmune diseases; (2) preoperative acceptance of any anti-tumor treatment; (3) a history of other malignant tumors; (4) incomplete clinical data; or(5) patients lost to follow-up.
Clinical Data Collection
The clinical data were collected as follows: hematological characteristics were obtained within one week before surgery. These date included neutrophil counts, lymphocyte counts, monocyte counts, and measurements of hemoglobin, platelets, albumin, lactate dehydrogenase, CEA, and CA199. Other clinical pathological data included gender, age, tumor location, tumor diameter, degree of differentiation, venous invasion, perineural invasion, and T and N stages. In addition, we staged all patients according to the seventh edition of the TNM staging system.4
Follow-Ups
Patients were followed up regularly according to the National Comprehensive Cancer Network (NCCN) guidelines. All patients were followed up every 3–6 months for the first two years, every six months within the third to fifth years, and then annually thereafter. Follow-up assessments included physical examinations and radiographic, colonoscopy and serologic tests. The last follow-up time was on August 1, 2019. The endpoint was the overall survival rate (OS). OS was defined as the time from surgery to death. In the training cohort, the median follow-up time was 65 months, and the number of OS events was 82(23.10%) at the last follow-up. In the validation cohort, the median follow-up time was 63 months, and the last follow-up OS event was 30(19.61%).
Statistical Analysis
Statistical analysis was performed using SPSS 23.0 (IBM, Chicago, IL, USA) and R software (version 3.1.4; http://www.Rproject.org). In addition to CEA, CA199, age, and tumor diameter, the best cutoff for all hematological biomarkers was obtained by X-tile 3.6.1 software (Yale University, New Haven, CT, USA). The categorical variables are expressed in terms of frequency and percentage and were compared using Chi-squared tests or Fisher exact tests.
First, all patients were randomly divided into a training cohort and a validation cohort in a ratio of 7:3. In the training cohort, a Cox regression model was used to analyze the risk factors by univariate and multi-factor analyses. Variables with significant factors in univariate COX analysis (P < 0.05) were included in multivariate analysis, and progressive risk factors were determined by forward stepwise selection. Then, we constructed a nomogram showing the three-year and five-year survival rates based on multivariate COX analysis results (using the rms package in R). In the training cohort and the verification cohort, the prediction accuracy and discriminating ability of the nomogram were evaluated, and Harrell’s C-index (C-index)and calibration curves were used. The maximum value of the C-index was 1.0, indicating that the predicted probability perfectly matched the actual probability, and 0.5 represented a random chance that the model correctly predicted the result. The nomogram and TNM staging systems were compared using decision curves. Finally, the total scores of each patient were calculated according to the established Cox regression model. Three groups of different prognostic risk subgroups (high risk, medium risk, and low risk) were classified by the X-tile procedure.12 The patient results (based on the total score) were constructed using the Kaplan Meier method, and the risk subgroups were classified as factors and compared by Log rank test. All statistical tests were two-sided, and P values less than 0.05 were statistically significant.
Ethical Statement
This retrospective study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University, and the study was conducted in accordance with the Declaration of Helsinki. Because this study was based on reviewing data from anonymous patients and did not involve patient intervention or the use of human tissue samples, no informed consent was required.
Results
Basic Characteristics
A total of 508 patients were enrolled in the study, including 355 patients in the training cohort and 153 patients in the validation cohort. The baseline characteristics of each group are shown in Table 1. In the training cohort, 170 patients (47.9%) were over 60 years old, 225 (63.4%) were males, and171 (48.2%) patients were confirmed to have colon cancer. Among them, 89 cases (25.1%) were in the T1/T2 stage, 85 cases (23.9%) were in the T3 stage, 172 cases (48.5%) were in the T4 stage, 220 cases (62.0%) were in the N0 stage, 81 cases (22.8%) were in the N1 stage, and 54 cases (15.2%) were in the N2 stage. The median follow-up time was 65 months. The three-year and five-year survival rates were 0.845 and 0.767, respectively. We found that 82 patients (23.10%) had died at the last follow-up. In the validation cohort, the distribution of these features was almost the same as that in the training cohort.
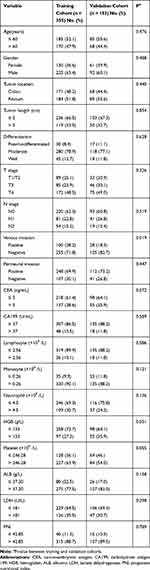 |
Table 1 Baseline Clinical Characteristics |
Univariate and Multivariate Analysis in the Training Cohort
All available information, including clinicopathological features and hematological biomarkers, were analyzed by univariate and multivariate Cox regression analysis of date from the 355 patients in the training cohort (Table 2). In the univariate analysis, there was a significant correlation between age, degree of differentiation, T stage, N stage, venous invasion, perineural invasion, CEA, CA199, neutrophils, monocytes, HGB, LDH, and PNI. Multivariate analysis was then performed to determine the factors that were distinguished in the univariate analysis. The results showed that age, differentiation, T stage, N stage, perineural invasion, neutrophils, monocytes, HGB, and LDH were independent risk factors for prognosis after radical resection of colorectal cancer.
 |
Table 2 Univariate and Multivariate Cox Hazards Analysis of Overall Survival in the Training Cohort |
Development of the Nomograms
Independent risk factors for OS were determined according to multivariate Cox regression analysis, and a nomogram was constructed to predict the three-year and five-year OS (Figure 1). In the training cohort, the C-index was 0.796 (95% CI: 0.761–0.831), which was higher than that of the TNM staging system (0.644, 95% CI: 0.595–0.694, P < 0.001). The calibration curves of the three-year and five-year OS were well matched to that of the standard line (Figure 2A and B).
 |
Figure 1 Nomograms for predicting overall survival. Abbreviations: HGB, Hemoglobin; LDH, lactate dehydrogenase. |
 |
Figure 2 The calibration curves for predicting the three- and five-year overall survival in the training (A, B) and validation (C, D) cohorts. |
Validation of the Nomograms
To further evaluate the predictive power, internal verification was performed in the verification cohort. The C index was 0.671 (95% CI: 0.656–0.686),which was better than that of the TNM staging system (0.665, 95% CI: 0.584–0.746). The calibration curves of the three-year and five-year OS were well matched to that of the standard line (Figure 2C and D).
Decision Curve Analysis
After determining the accuracy and discriminative ability of the model, we performed clinical validation on the nomogram through the validation cohort. The results showed that the nomogram has a good clinical applicability in predicting the survival of CRC because of its wide range of threshold probabilities (Figure 3). In addition, the nomogram had an advantage over traditional TNM staging systems in predicting OS because the net benefit was higher.
 |
Figure 3 Decision curve analysis for overall survival. Black line: All patients dead. Gray line: No patients died. Red line: Model of nomogram. Green line: Model of TNM staging system. |
Risk Stratification of OS
Patients were classified as low- (0–159 points), medium- (160–219 points), and high-risk subgroups (220 points or more) according to their nomogram scores. In the training cohort, there were 209 patients in the low-risk group, 70 patients in the intermediate-risk group, and 58 patients in the high-risk group. In the validation cohort, there were 83 patients in the low-risk group, 43 patients in the intermediate-risk group, and 27 patients in the high-risk group. There was a significant difference in the incidence of OS between the subgroups, and the survival rate of the high-risk subgroup was lower that than in the other groups(P < 0.05) (Figure 4).
Discussion
In this study, a nomogram model for predicting OS in patients undergoing radical resection of CRC was successfully established in combination with hematological biomarkers and clinical features, and was further verified in the validation cohort. This model had better predictive performance than that of the seventh edition of the AJCC TNM staging system.
In this study, we found hematological biomarkers including neutrophils, monocytes, HGB, and LDH to be independent risk factors for radical resection of CRC. Tumor-associated inflammation is one of the key features of cancer. In the case of body damage or pathogen invasion, the local immune system activates and induces the production of a large number of inflammatory cells (such as macrophages, mast cells, neutrophils, lymphocytes and other cells). In the tumor microenvironment, inflammatory cells can secrete a variety of cytokines, chemokines and cytotoxic mediators. These cells also induce cell carcinogenesis, and promote tumor cell infiltration and metastasis.13 Neutrophils are the first responding cells of the inflammatory response. They can promote tumor growth and induce adhesion and dissemination to distant organs by secreting various cytokines, including matrix metalloproteinase,14 chemokines15 and vascular endothelial growth factor (VEGF).16 In the early stages of colorectal tumors, neutrophil infiltration is also involved, because neutrophils infiltrate in colorectal adenomas much more than in adjacent normal mucosa. The number of neutrophils is positively correlated with the size of the adenoma.17 At the same time, lymphocytes are also one of the main participants in the inflammatory response, but their mechanism of action is still unclear. Previous studies have confirmed a correlation with the prognosis of a variety of tumors, including colorectal cancer.18 Studies have shown that anemia is also a risk factor for poor prognosis of cancer, and low levels of hemoglobin are associated with decreased patient survival.19 Hypoxia seems to be a factor influencing many types of cancer. Hypoxia can promote changes in tumor cells, thereby further prolonging the survival and malignant progression of tumors. Anemia is the main cause of tumor hypoxia.20 Our results also suggest that low levels of hemoglobin are associated with decreased survival in patients with colorectal cancer. Therefore, HGB seems to affect the survival of tumors. Studies have shown that elevated levels of LDH are associated with poor prognosis in various tumors.21 LDH has an inflammatory effect on the tumor microenvironment, activates interleukin (IL)-23 and IL-17, regulates the activity of arginase I, inhibits the activation of CD8+ T lymphocytes and natural killer (NK) cells, and helps cancer cells. evade the immune system.22 High levels of LDH promote tumor progression by inhibiting HIF-1 degradation and increasing vascular endothelial growth factor (VEGF) production.23 Several recent studies have also shown that LDH is a prognostic risk factor for CRC,24,25 which is consistent with the results of our present study.
Previous studies have also shown that the clinical characteristics of patients are also factors in CRC prognosis.8,26 In the present study, multivariate analysis found that age, T stage, N stage, differentiation and perineural invasion were independent risk factors for radical resection of CRC. Compared with the seventh edition of the AJCC TNM staging, our nomogram model exhibited better predictive performance. Zhang et al developed a nomogram containing the biomarker CEA to predict the OS of surgically resected CRC patients with a C-index higher than that of the seventh edition of the AJCC TNM staging system (0.710 vs 0.580).27 Fan et al Analyzed 13435 CRC patients and established a nomogram that included preoperative carcinoembryonic antigens, pT stages, negative lymph node counts, lymph node ratios (mLNR)and metastasis. The results indicated that the nomogram outperformed the AJCC stages with increased accuracy, net benefits and risk-assessment ability.28 The advantages of our present study include not only hematological biomarkers and clinical features, but also individualized predictions for patients. Due to the simple and graphical representation of the nomogram statistical prediction model, nomograms are usually based on the weight of the independent variables. Nomograms simultaneously integrate multiple independent variables in an easy-to-operate manner to predict the probability of clinical events numerically. In addition, in our present study, we divided patients into high, medium and low OS risk subgroups according to the nomogram total scores. There were significant differences in OS survival rates among the risk subgroups. Therefore, the results show that nomograms had a strong discriminating ability, and that patients with different risks can be stratified separately to provide patients with personalized treatment strategies and follow-up programs.
Although our nomogram provides a useful reference tool for clinicians, our research has several limitations. First, because this is a single-center retrospective study, potential biases cannot be ruled out. Second, our study did not evaluate the prognostic value of disease-free survival (DFS) in patients with CRC, and the C-index decay in the validation cohort is obvious, which may be due to the small sample size. Third, we had an insufficient sample size that prevented us from reaching more informative conclusions, and we only used internal verifications. Hence, in our future studies, we need to increase our sample sizes and incorporate external verifications to evaluate the applicability of nomogram to external populations.
In summary, we successfully established and validated a novel nomogram. This nomogram combined hematological biomarkers and clinical features to predict OS in patients undergoing radical resection of CRC, and it may provide a reference tool for clinicians to guide CRC patients with personalized treatments and follow-ups.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (81560726) and the Central Leading Local Science and Technology Development Special (ZY1949017).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Fidler MM, Soerjomataram I, Bray F. A global view on cancer incidence and national levels of the human development index. Int J Cancer. 2016;139(11):2436–2446. doi:10.1002/ijc.30382
4. Cuccurullo V, Mansi L. AJCC cancer staging handbook: from the AJCC cancer staging manual (7th edition). Eur J Nucl Med Mol Imaging. 2011;38(2):408.
5. Sagaert X, Vanstapel A, Verbeek S. Tumor heterogeneity in colorectal cancer: what do we know so far? Pathobiology. 2018;85(1–2):72–84. doi:10.1159/000486721
6. Tanio A, Saito H, Uejima C, et al. A prognostic index for colorectal cancer based on preoperative absolute lymphocyte, monocyte, and neutrophil counts. Surg Today. 2019;49(3):245–253. doi:10.1007/s00595-018-1728-6
7. Jiang H, Li H, Li A, et al. Preoperative combined hemoglobin, albumin, lymphocyte and platelet levels predict survival in patients with locally advanced colorectal cancer. Oncotarget. 2016;7(44):72076–72083. doi:10.18632/oncotarget.v7i44
8. Li Y, Jia H, Yu W, et al. Nomograms for predicting prognostic value of inflammatory biomarkers in colorectal cancer patients after radical resection. Int J Cancer. 2016;139(1):220–231. doi:10.1002/ijc.30071
9. Wang Y, Sun K, Shen J, et al. Novel prognostic nomograms based on inflammation-related markers for patients with hepatocellular carcinoma underwent hepatectomy. Cancer Res Treat. 2019;51(4):1464–1478. doi:10.4143/crt.2018.657
10. Li J, Chen S, Peng S, et al. Prognostic nomogram for patients with nasopharyngeal carcinoma incorporating hematological biomarkers and clinical characteristics. Int J Biol Sci. 2018;14(5):549–556. doi:10.7150/ijbs.24374
11. Ma M, Wang J, Hu Y, Weng M, Liu X, Wang Y. Prognostic value of inflammatory biomarkers in gastric cancer patients and the construction of a predictive model. Dig Surg. 2019;36(5):433–442. doi:10.1159/000493432
12. Zhang L-L, Li Y-Y, Hu J, et al. Proposal of a pretreatment Nomogram for predicting local recurrence after intensity-modulated radiation therapy in T4 nasopharyngeal carcinoma: a retrospective review of 415 Chinese patients. Cancer Res Treat. 2018;50(4):1084–1095. doi:10.4143/crt.2017.359
13. Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Curr Opin Genet Dev. 2008;18(1):3–10. doi:10.1016/j.gde.2008.01.003
14. Bausch D, Pausch T, Krauss T, et al. Neutrophil granulocyte derived MMP-9 is a VEGF independent functional component of the angiogenic switch in pancreatic ductal adenocarcinoma. Angiogenesis. 2011;14(3):235–243. doi:10.1007/s10456-011-9207-3
15. Tecchio C, Cassatella MA. Neutrophil-derived chemokines on the road to immunity. Semin Immunol. 2016;28(2):119–128. doi:10.1016/j.smim.2016.04.003
16. Tan KW, Chong SZ, Wong FHS, et al. Neutrophils contribute to inflammatory lymphangiogenesis by increasing VEGF-A bioavailability and secreting VEGF-D. Blood. 2013;122(22):3666–3677. doi:10.1182/blood-2012-11-466532
17. McLean MH, Murray GI, Stewart KN, et al. The inflammatory microenvironment in colorectal neoplasia. PLoS One. 2011;6:1. doi:10.1371/journal.pone.0015366
18. Yamamoto M, Saito H, Uejima C, et al. Combined pre- and postoperative lymphocyte count accurately predicts outcomes of patients with colorectal cancer. Dig Surg. 2019;36(6):487–494. doi:10.1159/000492340
19. Watine J, Bouarioua N. Anemia as an independent prognostic factor for survival in patients with cancer. Cancer. 2002;94(10):2793–2796. doi:10.1002/(ISSN)1097-0142
20. Boogaerts M, Mittelman M, Vaupel P. Beyond anaemia management: evolving role of erythropoietin therapy in neurological disorders, multiple myeloma and tumour hypoxia models. Oncology. 2005;69:22–30. doi:10.1159/000088285
21. Jurisic V, Radenkovic S, Konjevic G. The actual role of LDH as tumor marker, biochemical and clinical aspects. In: Scatena R, editor. Advances in Cancer Biomarkers: From Biochemistry to Clinic for a Critical Revision. Dordrecht:Springer; Vol. 867. 2015:115–124.
22. Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. Lactate dehydrogenase A: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018;7(12):6124–6136. doi:10.1002/cam4.2018.7.issue-12
23. Apicella M, Giannoni E, Fiore S, et al. Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 2018;28(6):848. doi:10.1016/j.cmet.2018.08.006
24. Guo G, Chen X, He W, et al. Establishment of inflammation biomarkers-based nomograms to predict prognosis of advanced colorectal cancer patients based on real world data. PLoS One. 2018;13:12. doi:10.1371/journal.pone.0208547
25. Roseweir AK, Clark J, McSorley ST, et al. The association between markers of tumour cell metabolism, the tumour microenvironment and outcomes in patients with colorectal cancer. Int J Cancer. 2019;144(9):2320–2329. doi:10.1002/ijc.v144.9
26. Jiang H, Tang E, Xu D, et al. Development and validation of nomograms for predicting survival in patients with non-metastatic colorectal cancer. Oncotarget. 2017;8(18):29857–29864. doi:10.18632/oncotarget.16167
27. Zhang Z-Y, Gao W, Luo Q-F, et al. A nomogram improves AJCC stages for colorectal cancers by introducing CEA, modified lymph node ratio and negative lymph node count. Sci Rep. 2016;6:39028.
28. Fan S, Li T, Zhou P, Peng Q, Zhu Y. Development and validation of nomogram combining serum biomarker for predicting survival in patients with resected rectal cancer. Biosci Rep. 2019;39(11).
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

