Back to Journals » Journal of Hepatocellular Carcinoma » Volume 8
Prognostic Model for the Risk Stratification of Early and Late Recurrence in Hepatitis B Virus-Related Small Hepatocellular Carcinoma Patients with Global Histone Modifications
Authors Duan JL , Nie RC , Xiang ZC, Chen JW, Deng MH, Liang H, Wang FW, Luo RZ, Xie D, Cai MY
Received 5 March 2021
Accepted for publication 6 May 2021
Published 28 May 2021 Volume 2021:8 Pages 493—505
DOI https://doi.org/10.2147/JHC.S309451
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmed Kaseb
Jin-Ling Duan,1,* Run-Cong Nie,1,2,* Zhi-Cheng Xiang,1,3,* Jie-Wei Chen,3 Min-Hua Deng,1,2 Hu Liang,1,4 Feng-Wei Wang,1 Rong-Zhen Luo,3 Dan Xie,1,3 Mu-Yan Cai1,3
1State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-Sen University Cancer Center, Guangzhou, 510060, People’s Republic of China; 2Department of Surgery, Sun Yat-Sen University Cancer Center, Guangzhou, 510060, People’s Republic of China; 3Department of Pathology, Sun Yat-Sen University Cancer Center, Guangzhou, 510060, People’s Republic of China; 4Department of Nasopharyngeal Carcinoma, Sun Yat-Sen University Cancer Center, Guangzhou, 510060, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Mu-Yan Cai; Dan Xie
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-Sen University Cancer Center, Guangzhou, 510060, People’s Republic of China
Tel +86-20-87343193
Fax +86-20-87343170
Email [email protected]; [email protected]
Background and Aim: To assess the profile of global histone modifications in small hepatocellular carcinoma (small HCC) and identify its prognostic value in predicting recurrence.
Methods: The expression profiles of global histone modifications, including H2AK5AC, H2BK20AC, H3K4me2, H3K9AC, H3K18AC, H4K12AC, and H4R3me2, were evaluated with immunohistochemistry in 335 HBV related small HCC patients. Two histone signature classifiers were then developed using least absolute shrinkage and selection operator Cox regression. A nomogram was built using the classifier and independent risk factors. The performances of the classifier and nomogram were assessed by receiver operating characteristic curves.
Results: Histone modifications were more pronounced in tumor tissues than in adjacent liver tissues. In tumor tissues, the risk score built based on the seven-histone signature exhibited satisfactory prediction efficiency, with an AUC = 0.71 (0.63– 0.79) for 2-year survival in the training cohort. Patients with a high risk score had shorter recurrence-free survival than those with a low risk score (HR: 1.96, 95% CI: 1.24– 3.08, p = 0.004; HR: 1.95, 95% CI: 1.12– 3.42, p = 0.019; and HR: 1.97, 95% CI: 1.39– 2.80, p < 0.001 for the training, validation and total cohorts, respectively). Furthermore, the statistical nomogram built using the histone classifier for early recurrence had a C-index = 0.68. In non-neoplastic liver tissues, the hepatic signature based on H3K4me2 and H4R3me2 was related to late recurrence (HR: 2.00, 95% CI: 1.15– 3.48, p = 0.01).
Conclusion: Global histone modifications in tumor and adjacent liver tissues are novel predictors of early and late recurrence, respectively, in HBV-related small HCC patients.
Keywords: small hepatocellular carcinoma, histone modifications, recurrence, LASSO, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-related death worldwide, with an incidence of approximately 850,000 new cases per year.1,2 Chronic HBV and HCV infections are the most important causes of HCC and account for 80% of HCC cases globally.3 Over the past decades, the surveillance of at-risk populations along with the development of imaging techniques has contributed to the stably increasing detection of early-stage HCC, particularly small HCC (≤ 3 cm).4 The diagnosis of small HCC allows patients to undergo potentially curative treatments, such as surgical resection and ablation.5 Unfortunately, the surgical outcomes of small HCC patients remain unsatisfactory because of the high rates of tumor recurrence, including early recurrence (≤ 2 years after resection) and late recurrence (> 2 years after resection). At present, for small HCC, 25–30% of patients experience early recurrence,6–8 and the 10-year recurrence-free survival (RFS) rate is only 22%, which is the main contributor to the 35% 10-year overall survival rate.9 Therefore, it is necessary to identify small HCC patients with a high risk of recurrence to provide earlier intervention.
Over the last two decades, various prognostic systems have been proposed to address the inter-relationship of prognostic factors among HCC patients and the complexity of HCC. The Barcelona Clinic Liver Cancer (BCLC) staging system is a crucial prognostic system for HCC patients and is widely used in clinical practice. However, the outcomes vary within the same stage, suggesting that the current systems, which mainly depend on clinicopathologic factors, inadequately reflect the biological heterogeneity of HCC, particularly small HCC. Thus, the development of a novel comprehensive prognostic system that combines biomarkers for clinical practice is crucial.
Tumors are not only genetic diseases but also epigenetic diseases.10 Epigenetic alterations in cancer comprise genome-wide and locus-specific changes in DNA methylation and posttranslational histone modifications, which influence chromatin accessibility and gene activity without changing the DNA sequence.11,12 The reversible posttranslational modifications of histones, such as lysine acetylation, lysine methylation, and arginine methylation, are modified by chromatin-modifying enzymes. Previous studies have shown that overexpression of the writer EZH2, the protein that catalyzes repressive histone H3 lysine 27 trimethylation (H3K27me3), is significantly related to an aggressive biology and unfavorable prognosis in HCC.13–16 Global histone modifications have been a focus on biomarker development as effective predictors of clinical prognosis and treatment strategies in prostate, intestinal, pancreatic, and lung cancers.17–20 Seligson et al first reported that, in combination with histone 3 lysine 18 acetylation (H3K18AC), histone 3 lysine 9 acetylation (H3K9AC), histone 3 lysine 4 dimethylation (H3K4me2), histone 4 lysine 12 acetylation (H4K12AC) and histone 4 arginine 3 dimethylation (H4R3me2), these changes indicated the risk of tumor recurrence in patients with low-grade prostate cancer.17 Multiple histones, H3K4me2, histone 2A lysine 5 acetylation (H2AK5AC), histone 2B lysine 12 acetylation (H2BK12AC), H3K9AC, and histone 4 lysine 8 acetylation (H4K8AC), were further analyzed by Barlési et al, who found that evaluating selected changes may help to select early-stage NSCLC patients for adjuvant treatment.18 However, the implication of these global histone modifications in HCC, particularly HBV related small HCC, is poorly understood.
In the present study, we used immunohistochemistry to evaluate the levels of the abovementioned factors (H2AK5AC, H3K4me2, H3K9AC, H3K18AC, H4K12AC, and H4R3me2) in addition to histone 2B lysine 20 acetylation (H2BK20AC) to clarify the prognostic value of global histone modifications in predicting recurrence in HBV related small HCC patients and to establish an effective prognostic classifier for predicting relapse with a practical and quantitative nomogram.
Methods
Study Population
The Institutional Medical Ethics Committee of Sun Yat-Sen University Cancer Center (SYSUCC) approved the present retrospective study (approval number: YB2017-026) and the study was informed in accordance with Declaration of Helsinki. Written informed consent was obtained from the patients before the study began. Pathologically confirmed, nonmetastatic small HCC samples were collected from the SYSUCC between 1998 and 2010. The inclusion criteria were as follows: (1) solitary small HCC (≤3 cm); (2) the presence of HBV surface antigens; (3) primary and curative resection; (4) no metastatic or residual disease; (5) no preoperative adjuvant therapy; and (6) complete clinical information and follow-up data. All the cases included in this study were diagnosed by experienced pathologists. Tumor grades were determined following the criteria of the Edmondson-Steiner grading system. Patients with unknown causes of death were excluded.
Follow-Up and Outcomes
The evaluation and management approaches employed before surgical resection were previously described. After curative partial hepatectomy, the patients were examined using abdominal ultrasonography, chest radiography, laboratory tests and serum AFP analysis. These assessments were performed 1 month after resection and then generally in 3-month intervals in the first 2 years and every 3–6 months in subsequent years until tumor recurrence was documented. For patients in whom tumor recurrence was suspected, further tests, including computed tomography (CT) and magnetic resonance imaging (MRI), were performed, and biopsies were employed if necessary. Patients with confirmed recurrence underwent appropriate further treatment.
RFS was defined as the time from the date of surgery to the date of first tumor recurrence (local or distant metastasis identified by imaging and pathology). Cancer-specific overall survival (OS) was defined as the time from the date of surgery to the date of death as a result of HCC.
Tissue Microarray and Immunohistochemistry
Tissue microarrays (TMAs) were assembled in accordance with our previous methodology.21 Triplicate 0.6-mm diameter cylindrical areas were punched from representative regions of an individual donor tissue block and re-embedded into a recipient paraffin block in a defined position using a tissue arraying instrument (Beecher Instruments, Silver Spring, MD).
A standard, two-step, indirect immunohistochemical staining method was used as previously described.14 The histone modification markers employed in this report were selected based on previous studies showing that the modifications were applicable in other carcinomas.17,18 The following primary antibodies were applied for 60 minutes at room temperature: rabbit anti-histone polyclonal antibodies against H2AK5AC, H2BK20AC, H3K4me2, H3K9AC, H3K18AC, and H4K12AC (Cell Signaling Technology, Beverly, MA, USA; 1:100 dilution) and a rabbit monoclonal anti-H4R3me2 antibody (Abcam, Cambridge, MA; 1:200 dilution). Positive and negative control samples were included on each slide. All antibody-related information is shown in Table S1. Semiquantitative assessments for histone modifications were scored by the percentage of tumor cells with positive nuclear staining over the total number of tumor cells from three cylinders (0.6 mm diameter/cylinder) of each case. Scores were assigned by using 5% increments (0%, 5%, 10% … 100%). The evaluations were performed independently by two experienced pathologists (M-Y Cai and R.-Z. Luo), who were blinded to all clinicopathologic and outcome variables. Individual scores were averaged; in cases of a difference of more than 20%, a third pathologist (D Xie) was consulted and provided a score. The final score was determined by the average based on the assessments of the three pathologists.
Histone Modification Risk-Score Classifier and Nomogram
A stratified randomization approach via the “resample” package was used to divide the small HCC dataset into training and validation cohorts at a ratio of 6:4 to construct and validate the histone risk score. After overviewing the density distribution of each histone modification, further relative stratification into high expression and low expression revealed two subgroups of patients within each modification. The cut-off value of each histone modification marker to predict survival was determined with the “survminer” package. The cut-off scores generated with the training cohort were applied to the validation and total cohorts.
The Cox proportional hazards model with LASSO regression (“glmnet” package) was applied to construct the histone risk score, which was broadly applied for high-dimensional and sample-limited data.22,23 The optimal values of the penalty parameter λ were determined by 200-fold cross-validation using the minimum criteria on the training dataset. Time-dependent receiver operating characteristic (tdROC) curves and areas under the curves (AUCs) were generated using the “survivalROC” package to assess prognostic accuracy.24 Patients were grouped into high-risk-score and low-risk-score groups based on the cut-off, which was the median value of the training cohort.
The visual nomogram for recurrence was formulated by using the independent risk factors identified in the multivariate Cox regression analysis. Calibration was evaluated with the calibration plot. The discrimination ability of the nomogram was assessed by tdROC analysis. Moreover, decision curve analysis (DCA) with the stdca function was performed to evaluate the improvement in clinical decision making.
Statistical Analysis
One-way ANOVA and chi-square tests were used to assess differences in continuous and categorical variables, respectively. Pearson’s correlation test was applied to quantify linear correlations among the histone modifications. The Kaplan–Meier method with the Log rank test was employed to estimate the survival rates. The Cox regression model was utilized for multivariate survival analysis. All statistical analyses were performed using R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria, http://www.rproject.org). All statistical tests were two-sided, and p values less than 0.05 were considered significant. All data in our study have been recorded at Sun Yat-sen University Cancer Center with a Research Data Deposit (RDD) number of RDDB2021001160.
Results
Characteristics of the Small HCC Cohort and Global Histone Modifications
The baseline characteristics of the small HCC patients in this study are summarized in Table 1. A total of 335 patients were enrolled in our study, including 295 males and 40 females, with a median age of 49 years (range, 26–78 years). With a median follow-up time of 49 months (interquartile range, 36–67 months), 131 (39.1%) patients developed recurrence, and 62 (18.5%) died. The 1-, 2-, and 5-year OS and RFS rates in the total cohort were 97.9%, 93.9% and 77.7%, and 89.3%, 76.4% and 59.9%, respectively.
 |
Table 1 Characteristics of Small HCC Patients in the Training, Validation and Total Cohorts |
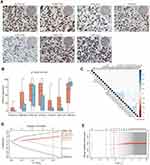
To determine the global levels of histone modifications, we referred to previous studies and selected H2AK5AC, H2BK20AC, H3K4me2, H3K9AC, H3K18AC, H4K12AC and H4R3me2 to further investigate in small HCC. Both primary small HCC and paired non-neoplastic liver tissues were stained with specific antibodies. Semiquantitative assessments of the percentage of tumor cells with positive nuclear staining over total tumor cells (range, 0% to 100% using 5% increments) in three fields were performed, and individual scores were averaged. Representative images of staining for these markers showed clear and distinguishable brown staining of histones in the nucleus (Figure 1A), and the density distribution of each histone modification is shown in Figure S1. Compared to those in paired non-neoplastic liver tissues, all modifications except for H4R3me2 showed significantly stronger staining in tumor tissues (Figure 1B, all for p < 0.01). Interestingly, different histone modifications were found to correlate within tumor (r, 0.16–0.56) or non-neoplastic liver tissues (r, 0.19–0.51), which might be consistent with the existence of combinatorial patterns of histone modifications.25 However, only weak correlations were found between clinicopathologic characteristics and histone modifications (Figure 1C).
Risk Score Classifier Based on Global Histone Modifications in Small HCC
To eliminate collinearity among factors, Cox regression with the LASSO model was performed to construct a histone modification classifier: the histone risk score (Figure 1D and E). The formula of the classifier was derived based on the histone modification features in the training cohort with small HCC, and seven modifications were included after filtering. The histone risk score was generated as follows: (0.195 × scales of H2AK5AC) + (0.167 × scales of H2BK20AC) – (0.733 × scales of H3K4me2) + (0.086 × scales of H3K9AC) + (0.155 × scales of H3K18AC) + (0.403 × scales of H4K12AC) – (0.298 × scales of H4R3me2).
To assess the predictive value of the histone risk score, a tdROC curve was created. As shown in Figure 2A, the AUCs of the risk score in the training cohort at 1 year, 2 years and 3 years were 0.65 (0.52–0.77), 0.71 (0.63–0.79) and 0.68 (0.60–0.77), respectively. The AUCs in the validation cohort were 0.69 (0.57–0.81), 0.58 (0.5–0.68) and 0.63 (0.52–0.73), and those in the total cohort were 0.67 (0.58–0.75), 0.66 (0.59–0.72) and 0.66 (0.60–0.73), respectively.
High- and low-risk-score groups were divided by the median risk score from the training cohort, and the distribution of clinical characteristics did not vary significantly between the two subgroups (Table S2). In the training cohort, the high-risk group showed a worse RFS than the low-risk group (HR: 1.96; 95% CI: 1.24–3.08; p = 0.004; Figure 2B). Similar results were obtained in the validation cohort (HR: 1.95; 95% CI: 1.12–3.42; p = 0.019; Figure 2B) and the total cohort (HR: 1.97; 95% CI: 1.39–2.80; p < 0.001; Figure 2B). Additionally, a high risk score indicated significantly worse overall survival in the three cohorts (Figure S2).
Intriguingly, compared with the low-score group, the high-score group had more early recurrences (Figure 3A). The two-year RFS rates of the high-score group and low-score group were 84.5% vs 67.2% (p = 0.001) in the training cohort, 79.6% vs 71.9% in the validation cohort (p = 0.21), and 82.7% vs 69.2% in the total cohort (p < 0.001) (Figure 3B). Furthermore, the histone risk score of those with early recurrence was evidently higher than that of those without recurrence (Figure S3A; p < 0.01 for the total cohort). The prognostic value of the classifier for early recurrence was evaluated by ROC curve analysis (Figure S3B). These results indicated that the histone risk score could effectively predict recurrence and even early recurrence in patients with small HCC.
Histone Modification Risk Score as an Independent Prognostic Factor
In the unadjusted analysis, the following were revealed as prognostic factors for poor RFS: advanced pathologic grade, tumor size larger than 2 cm, the presence of vascular invasion, tumor necrosis, and a high histone modification risk score (Table 2). The 1-, 2-, and 5-year RFS rates of the high- and low-risk groups were 84.6% vs 92.7%, 69.2% vs 82.7%, and 50.6% vs 68.0% (HR: 1.97; 95% CI: 1.39–2.80; p < 0.001).
 |
Table 2 Univariate and Multivariate Analyses for Recurrence-Free Survival |
The results of the multivariate Cox proportional hazards model are shown in Table 2. After adjusting for other factors, the analysis revealed that the histone risk score was an independent adverse prognostic risk factor for HBV related small HCC patients regarding RFS (HR: 1.95; 95% CI: 1.37–2.77; p < 0.001; Table 2), with similar results observed for OS (HR: 2.11; 95% CI: 1.22–3.63; p = 0.007; Table S3). An advanced pathologic grade (HR: 1.47; 95% CI: 1.15–1.88; p = 0.002), a tumor size larger than 2 cm (HR: 1.52; 95% CI: 1.07–2.16; p = 0.020) and tumor necrosis (HR: 1.79; 95% CI: 1.25–2.55; p = 0.001) were also revealed as independent predictors for small HCC recurrence.
Nomogram Construction
To visualize the early recurrence prediction model, a nomogram was built based on the histone risk score and individual significant clinicopathologic predictors (including tumor size, pathologic grade and tumor necrosis) (Figure 4A). The calibration curve showed favorable agreement between the prediction and actual observation (Figure 4B). The predictive accuracy of the nomogram was excellent, with a C-index = 0.68 and an AUC = 0.70 (0.63–0.76) for predicting early recurrence and 5-year overall survival (Figure 4C; Figure S2B). When the nomogram-derived merge score was used, for patients with high and low merge scores based on the median values, the 2-year RFS rates were 66.6% vs 85.5% (HR: 2.47; 95% CI: 1.72–3.55; p < 0.001) (Figure 4D). Additionally, decision curve analysis (DCA) was performed to evaluate the improvement in clinical decision making (Figure S4A).
Histone Modifications in Adjacent Non-Neoplastic Hepatic Tissues
To further explore the prognostic value of histone modifications and considering that the biological signature of adjacent non-neoplastic tissues tends to be associated with late recurrence, as reported by Hoshida et al, the LASSO model was also separately applied in adjacent non-neoplastic liver tissues (Figure S4B and C).26 Two histone modifications in non-neoplastic liver tissues, H3K4me2 and H4R3me2, were selected via the LASSO model and included in the hepatic score for late recurrence (AUC = 0.62; 95% CI: 0.53–0.70) (Figure S4D). Notably, the hepatic score based on these histone modifications in non-neoplastic liver tissues could significantly predict the survival of patients with late recurrence (HR: 2.00; 95% CI: 1.15–3.48; p = 0.01) (Figure 4E and F). In agreement with the study by Xu et al, our study demonstrated that patients with late recurrence had shorter overall survival times than those without recurrence (p < 0.001, data not shown).27 Our preliminary investigation first proposed and suggested that histone modifications in adjacent non-neoplastic liver tissues could predict late recurrence in HBV related small HCC.
Discussion
To our best knowledge, this study was the first to focus on HBV-associated small hepatocellular carcinoma to profile the global expression of histone modifications in tumor and nontumor liver tissues. We described the development and validation of a promising recurrent risk score signature based on a panel of histone modifications, including H2AK5AC, H2BK20AC, H3K4me2, H3K9AC, H3K18AC, H4K12AC and H4R3me2, which was identified as an independent prognostic risk factor for recurrence-free survival and overall survival. A novel prognostic nomogram was further established with a combination of the histone risk score and other significant traditional predictors, showing satisfactory predictive accuracy for recurrence in patients with HBV related small HCC. More interestingly, histone modifications in non-neoplastic liver tissue might be predictors for the late recurrence of small HCC. Our findings highlight a novel method with which the levels of histone modifications may be used to predicting the early and late recurrence of small HCC with HBV.
Due to the complexity and heterogeneity of HCC, it is complicated and challenging to identify and apply a single molecular profile for clinical prediction. Although precision diagnosis using gene expression profiling has been developed, its high cost and unsatisfactory reproducibility restrict its application. In contrast, histone modifications offer an understandable viewpoint as transcriptional regulators reflecting the overall perspective of cells, generally via chromatin modification to promote or repress target gene expression. However, the epigenome and its regulation and dysregulation in cancer are complex, and the molecular mechanism still needs further investigation. The sum of all different histone modifications determines the chromatin structure and biological outcomes.28 Previous studies have revealed that several global histone modifications are associated with prognosis in various cancers based on overall expression by IHC staining; however, related research is lacking in HCC.17,18,20,29–31 In the present study, we first assessed a panel of global histone modifications in HBV related small HCC and found that these histone modifications (except for H4R3me2) were more pronounced in tumor tissues than in paired non-neoplastic tissues. Studies have indicated that H2AK5AC, H3K9AC, H3K18AC, H4K12AC, and H3K4me2 are associated with transcriptionally active gene promoters, and H4R3me2 is associated with gene repression.29,32–36 The less studied H2BK20AC is a good predictor of active enhancers.37 Additionally, in contrast to previous studies, we noticed that different carcinomas might have different patterns of histone modifications; for example, approximately half of the small HCC samples had less than 30% H3K9AC staining; however, in prostate cancer, more than 50% of the samples showed 90–100% staining.17 Furthermore, consistent with Seligson et al’s study in lung and kidney cancer patients, we found that a low level of H3K4me2 in small HCC contributes to a poor prognosis, which might be attributed to decreased methylation levels at repetitive DNA elements and an aggressive phenotype.38
Recurrence is a primary issue that severely impacts the survival of HCC patients. In addition to the clinical staging system, much effort has been made to identify patients with a high risk of recurrence. Increasing evidence has revealed that early recurrence is mainly associated with the nature of the primary tumor, such as tumor size and the number of nodular and vascular invasions, while the above presentations often appear in advanced stages and not early stages, such as small HCC; thus, biological markers for small HCC are still needed.39–43 We reviewed the research over the last ten years, and few studies have focused on HBV related small HCC biomarkers.44–47 In the present study, we revealed that the panel of global histone modifications in tumor tissues could be a significantly strong independent risk factor for small HCC, and patients with higher scores tended to experience more relapses, showing that histone modifications effectively predict recurrence and survival.
Additionally, we presented results for the first time that histone modifications in non-neoplastic liver tissues are related to the late recurrence of small HCC. In contrast to that of early recurrence, the mechanism of late recurrence is far from being fully understood.26 Late recurrence likely arises from a de novo tumor and is more relevant to the characterization of the field of adjacent liver tissues; however, the underlying mechanism remains to be elucidated.26 The analysis of non-neoplastic liver tissues showed that the expression of a group of genes was related to late recurrence but not early recurrence.26,48 Additional studies proposed that methylation patterns have already changed in normal tissues before carcinogenesis is detected.19,49–51 However, to the best of our knowledge, none of the histone modifications in non-neoplastic tissues have been investigated. This study highlights that histone modification levels could serve as predictive biomarkers not only for early recurrence but also for late recurrence. To further improve the survival of HBV related small HCC patients, patients with different risks must be stratified so that those with a high risk for early recurrence can receive more aggressive treatment after resection, while patients with a high risk of late recurrence, and even those who achieve 2 years of recurrence-free survival, should still receive regular surveillance to improve survival.27
We acknowledge that limitations exist in the present study. First, the study was retrospective in design and occurred at a single institute, although the present study was based on a solitary HBV-associated small HCC cohort with complete clinical information. Due to the homogeneity of resected small HCC, the number of samples was relatively small, which might have led to the observed trends for early recurrence and no statistical significance in the validation cohort. However, a trend toward significance was observed in the validation cohort. Second, although internal validation was performed to reduce overfitting of the model and improve its robustness for prediction, only 7 biomarkers were included in the present study, and no variables were discarded in the LASSO model. More importantly, external validation is still needed to generalize the use of the predictive model before it can be applied to clinical practice in the real world. Third, this clinical study did not reveal how these specific epigenetic modifications or combinations of changes affect the expression of particular genes or tumor behavior, which remains unclear, and the mechanistic basis needs to be further explored.
Conclusion
The present study is the first to assess the profiles of global histone modifications in hepatocellular carcinoma. We identified a histone risk score based on a panel of global histone modifications in tumors that could act as an independent predictor of early recurrence and survival in small HCC patients with HBV. Furthermore, we found that histone modifications in adjacent non-neoplastic liver tissues may be associated with late recurrence. This system could be useful to stratify patients according to the risk of tumor recurrence.
Abbreviations
Small HCC, small hepatocellular carcinoma; TMA, tissue microarray; LASSO, least absolute shrinkage and selection operator; ROC, receiver operating characteristic; AUC, area under the curve; HR, hazard ratio; RFS, recurrence-free survival; BCLC staging, Barcelona Clinic Liver Cancer staging; AFP, alpha-fetoprotein; CT, computed tomography; MRI, magnetic resonance imaging; OS, overall survival; tdROC, time-dependent receiver operating characteristic; DCA, decision curve analysis.
Data Sharing Statement
The related data are available for sharing upon request to Pro. Mu-Yan Cai.
Ethics Approval and Consent to Participate
The Institutional Medical Ethics Committee of Sun Yat-Sen University Cancer Center (SYSUCC) approved this retrospective study and patients provided written informed consent (approval number: YB2017-026).
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (No. 2017YFC1309000), the National Natural Science Foundation of China (grant Nos. 81730072, 81672407, 81872001, and 82002855), the Guangzhou Science and Technology Plan Projects (Health Medical Collaborative Innovation Program of Guangzhou, No. 201803040019), the Guangdong Natural Science Funds for Distinguished Young Scholar (No. 2015A030306001), and China Postdoctoral Science Foundation (No. 2019M663293). The authors greatly thank Professor Xueqin Wang and Canhong Wen for statistical assistance during the preparation of this manuscript.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
1. Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018.
2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
3. Park JW, Chen M, Colombo M, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE study. Liver Int. 2015;35(9):2155–2166. doi:10.1111/liv.12818
4. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362(9399):1907–1917. doi:10.1016/S0140-6736(03)14964-1
5. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi:10.1002/ijc.29210
6. Huang J, Yan L, Cheng Z, et al. A randomized trial comparing radiofrequency ablation and surgical resection for HCC conforming to the Milan criteria. Ann Surg. 2010;252(6):903–912. doi:10.1097/SLA.0b013e3181efc656
7. Feng K, Yan J, Li X, et al. A randomized controlled trial of radiofrequency ablation and surgical resection in the treatment of small hepatocellular carcinoma. J Hepatol. 2012;57(4):794–802. doi:10.1016/j.jhep.2012.05.007
8. Roayaie S, Obeidat K, Sposito C, et al. Resection of hepatocellular cancer ≤2 cm: results from two Western centers. Hepatology (Baltimore, Md). 2013;57(4):1426–1435. doi:10.1002/hep.25832
9. Poon RT, Fan ST, Lo CM, Liu CL, Wong J. Long-term survival and pattern of recurrence after resection of small hepatocellular carcinoma in patients with preserved liver function: implications for a strategy of salvage transplantation. Ann Surg. 2002;235(3):373–382. doi:10.1097/00000658-200203000-00009
10. Huang J. Current progress in epigenetic research for hepatocarcinomagenesis. Sci China Series C. 2009;52(1):31–42.
11. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi:10.1038/nrg2005
12. Ting AH, McGarvey KM, Baylin SB. The cancer epigenome–components and functional correlates. Genes Dev. 2006;20(23):3215–3231. doi:10.1101/gad.1464906
13. He C, Xu J, Zhang J, et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Human Pathol. 2012.
14. Cai MY, Hou JH, Rao HL, et al. High expression of H3K27me3 in human hepatocellular carcinomas correlates closely with vascular invasion and predicts worse prognosis in patients. Mol Med. 2011;17(1–2):12–20. doi:10.2119/molmed.2010.00103
15. Zheng F, Liao YJ, Cai MY, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61(2):278–289. doi:10.1136/gut.2011.239145
16. Gao SB, Xu B, Ding LH, et al. The functional and mechanistic relatedness of EZH2 and menin in hepatocellular carcinoma. J Hepatol. 2014;61(4):832–839. doi:10.1016/j.jhep.2014.05.015
17. Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi:10.1038/nature03672
18. Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol. 2007;25(28):4358–4364. doi:10.1200/JCO.2007.11.2599
19. Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(5):1204–1225 e1212.
20. Manuyakorn A, Paulus R, Farrell J, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 2010;28(8):1358–1365. doi:10.1200/JCO.2009.24.5639
21. Cai MY, Tong ZT, Zheng F, et al. EZH2 protein: a promising immunomarker for the detection of hepatocellular carcinomas in liver needle biopsies. Gut. 2011;60(7):967–976. doi:10.1136/gut.2010.231993
22. Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16(4):385–395. doi:10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3
23. Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi:10.1016/S1470-2045(13)70491-1
24. Zou KH, Hall WJ, Shapiro DE. Smooth non-parametric receiver operating characteristic (ROC) curves for continuous diagnostic tests. Stat Med. 1997;16(19):2143–2156. doi:10.1002/(SICI)1097-0258(19971015)16:19<2143::AID-SIM655>3.0.CO;2-3
25. Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi:10.1038/ng.154
26. Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359(19):1995–2004. doi:10.1056/NEJMoa0804525
27. Xu XF, Xing H, Han J, et al. Risk factors, patterns, and outcomes of late recurrence after liver resection for hepatocellular carcinoma: a multicenter study from China. JAMA Surg. 2019;154(3):209–217. doi:10.1001/jamasurg.2018.4334
28. Audia J, Campbell R. Histone modifications and cancer. Cold Spring Harbor Perspect Biol. 2016;8(4):a019521. doi:10.1101/cshperspect.a019521
29. Elsheikh SE, Green AR, Rakha EA, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69(9):3802–3809. doi:10.1158/0008-5472.CAN-08-3907
30. Tzao C, Tung HJ, Jin JS, et al. Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod Pathol. 2009;22(2):252–260. doi:10.1038/modpathol.2008.172
31. Chen YW, Kao SY, Wang HJ, Yang MH. Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer Am Cancer Soc. 2013;119(24):4259–4267.
32. Vavouri T, Lehner B. Human genes with CpG island promoters have a distinct transcription-associated chromatin organization. Genome Biol. 2012;13(11):R110. doi:10.1186/gb-2012-13-11-r110
33. Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. doi:10.1038/ncb1076
34. Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi:10.1016/j.cell.2005.01.001
35. Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–527. doi:10.1016/j.cell.2005.06.026
36. Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinform. 2010;11:396. doi:10.1186/1471-2105-11-396
37. Kumar V, Rayan NA, Muratani M, et al. Comprehensive benchmarking reveals H2BK20 acetylation as a distinctive signature of cell-state-specific enhancers and promoters. Genome Res. 2016;26(5):612–623. doi:10.1101/gr.201038.115
38. Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174(5):1619–1628. doi:10.2353/ajpath.2009.080874
39. Chan AWH, Zhong J, Berhane S, et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018;69(6):1284–1293. doi:10.1016/j.jhep.2018.08.027
40. Liu PH, Hsu CY, Hsia CY, et al. Prognosis of hepatocellular carcinoma: assessment of eleven staging systems. J Hepatol. 2016;64(3):601–608. doi:10.1016/j.jhep.2015.10.029
41. Marasco G, Colecchia A, Colli A, et al. Role of liver and spleen stiffness in predicting the recurrence of hepatocellular carcinoma after resection. J Hepatol. 2019;70(3):440–448. doi:10.1016/j.jhep.2018.10.022
42. Wu JC, Huang YH, Chau GY, et al. Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J Hepatol. 2009;51(5):890–897. doi:10.1016/j.jhep.2009.07.009
43. Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg. 2015;261(5):947–955. doi:10.1097/SLA.0000000000000710
44. Liu Y, Jiang P, Wang G, Liu X, Luo S. Downregulation of RFX1 predicts poor prognosis of patients with small hepatocellular carcinoma. Eur J Surg Oncol. 2018;44(7):1087–1093. doi:10.1016/j.ejso.2018.04.017
45. Gu J, Zhang X, Miao R, et al. A three-long non-coding RNA-expression-based risk score system can better predict both overall and recurrence-free survival in patients with small hepatocellular carcinoma. Aging. 2018;10(7):1627–1639. doi:10.18632/aging.101497
46. Tan Y, Yin P, Tang L, et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol Cell Proteom. 2012;11(2):
47. Jin G, Li Y, Cong W, et al. iTRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J Proteome Res. 2011;10(8):3418–3428. doi:10.1021/pr200482t
48. Kim JH, Sohn BH, Lee HS, et al. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med. 2014;11(12):e1001770. doi:10.1371/journal.pmed.1001770
49. Yoshida S, Yamashita S, Niwa T, et al. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer. 2017;20(1):136–145. doi:10.1007/s10120-016-0593-5
50. Utsunomiya T, Shimada M, Morine Y, Tajima A, Imoto I. Specific molecular signatures of non-tumor liver tissue may predict a risk of hepatocarcinogenesis. Cancer Sci. 2014;105(7):749–754. doi:10.1111/cas.12431
51. Lou C, Du Z, Yang B, Gao Y, Wang Y, Fang S. Aberrant DNA methylation profile of hepatocellular carcinoma and surgically resected margin. Cancer Sci. 2009;100(6):996–1004. doi:10.1111/j.1349-7006.2009.01138.x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.