Back to Journals » OncoTargets and Therapy » Volume 13
Pristimerin Suppressed Breast Cancer Progression via miR-542-5p/DUB3 Axis
Authors Cheng S , Zhang Z, Hu C , Xing N , Xia Y, Pang B
Received 16 April 2020
Accepted for publication 16 June 2020
Published 7 July 2020 Volume 2020:13 Pages 6651—6660
DOI https://doi.org/10.2147/OTT.S257329
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
Shihuan Cheng,1,* Zhihong Zhang,2,* Cong Hu,2,3 Na Xing,4 Yan Xia,5 Bo Pang3,6
1Department of Rehabilitation, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China; 2Centre for Reproductive Medicine, Centre for Prenatal Diagnosis, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China; 3Central Laboratory, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China; 4Department of Pediatrics, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China; 5Department of Gastroenterology, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China; 6Department of Cardiology, The First Hospital of Jilin University, Changchun, Jilin 130021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yan Xia; Bo Pang Email [email protected]; [email protected]
Background: Breast cancer is one of the most common and malignant tumors in the world. Nowadays more attention has been garnered in pristimerin anti-cancer effects. Here, we illustrate the function and regulatory mechanism of pristimerin in breast cancer therapy.
Materials and Methods: Breast cancer cell lines MCF-7, MDA-MB-231, and 4T1 were used. Cell Counting Kit-8 (CCK-8) assay was performed to evaluate proliferation viability of breast cancer cells under pristimerin treatment. Wound healing assay was used to examine the migration ability, cell cycle, and cell apoptosis detection were tested by flow cytometry. Bioinformatic analysis was used to find the underlying molecular and gene connected with pristimerin and breast cancer survival. Finally, we used transfection and real-time polymerase chain reaction analysis to confirm the mechanism.
Results: We observed that pristimerin inhibited breast cancer cell viability, migration, and cell cycle, meanwhile induced cell apoptosis. In addition, under pristimerin treatment, miR-542-5p was up-regulated while DUB3 was down-regulated. Furthermore, bioinformatics analysis showed higher expression of DUB3 in breast cancer compared with normal tissue, also with poor prognosis. Overexpression miR-542-5p in breast cancer cells leads to a decrease in DUB3 level. The effect was obviously post pristimerin treatment and miR-542-5p overexpression.
Conclusion: Pristimerin inhibited breast cancer progression through DUB3 expression via a canonical miRNA-mediated mechanism.
Keywords: pristimerin, breast cancer, DUB3, AGO2, miRNA, anti-cancer
Introduction
Breast cancer is one of the most common primary tumors in females worldwide, also with a high malignancy degree.1 Over the past few years, technical advances, diagnosis criteria, and therapy strategies have improved, though therapeutic efficacy is still limited, and mortality rates of breast cancer have reduced.2,3 It’s crucial to improve the treatment and prognosis of patients.4
Pristimerin (20α-3-hydroxy-2-oxo-24-nor-friedela-1-10,3,5,7-tetraen-carboxylic acid-29-methyl ester) isolated from the Celastraceae and Hippocrateaceae families is a naturally occurring quinone methide triterpenoid. Various pharmacological effects of pristimerin have been confirmed, including insecticidal, anti-inflammatory, anti-angiogenic, anti-protozoal, and anti-cancer roles.5 Nowadays, the anti-cancer activity as a novel function of pristimerin has been discovered in cancer cell lines (eg, glioma, colon, and multiple myeloma), and research is focused on the various molecular targets and pathways of pristimerin in cancer treatment and prevention of chemotherapy resistance.6,11
MicroRNAs (miRNAs) are a large family of small non-coding RNAs containing 20–22 nucleotides.12 miRNAs target the RNA induced silencing complex (RISC) by binding to specific sites within the 3ʹ-UTR (three prime untranslated region) to decrease the expression of target mRNAs at the post-transcriptional level,13 leading to either mRNA degradation or the inhibition of protein translation.14 Argonaute (AGO) protein is an important member of RISC. Evidence has shown that miRNAs play a key role in a number of critical biological processes leading to a broad spectrum of diseases. Particularly, whether as tumor suppressor genes or oncogenes, miRNAs can coordinate multiple cellular processes related to cancer progression and disease severity,15 which means that miRNAs can not only be a crucial pathophysiological component associated with tumor development, but also a therapy target for cancer.
Ubiquitination is a reversible process and ubiquitin moieties are removed from polypeptides by deubiquitinases (DUBs, also named USP17L2). Based on accumulating evidence, DUBs are essential for DNA repair, transcription, and cell cycle progression and other cellular functions regulation.16 The DUBs family includes five subgroups, DUB3 is an immediate early gene which is also cytokine-inducible.17 It have been confirmed that DUB3 expression was related to tumor proliferative potential and apoptosis progression, leading to poor prognoses in various human cancers (eg, non-small cell lung cancer, epithelial ovarian cancer, and colorectal cancer).18,20 However, the role of DEB3 in mediating tumor cell migration and metastasis is still unclear.
Here, we utilize unbiased approaches to identify the specific pristimerin responsible for anti-cancer effect, and identify miR-542-5p as a silence signal of DUB3. The miR-542-5p/DUB3 signaling axis forms a “sensor and effector” circuitry by pristimerin treatment to suppress breast cancer progression.
Materials and Methods
Materials
Pristimerin (purity ≥ 99%) was purchased from Paypaytech Inc. (Shenzhen, China), reconstructed with dimethyl sulfoxide to 20 mM stock solution, and stored in small aliquots at −20°C.
Cells and Cell Culture
Breast cancer cell lines MCF-7, MDA-MB-231, and 4T1 were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA), glutamine (2 mmol/L), streptomycin (100 μg/mL), penicillin (100 U/mL), and were maintained at 37°C in humidified 5% CO2 incubators. Cells in the mid-log phase were used for further research. For cell count, medium was removed from the culture container, rinsed with PBS, then cells were exposed to trypsin, and incubated for 1–2 minutes. Next, trypsin activity was stopped by adding serum containing medium, pipetting the cells up and down until the cells were dispersed into a single cell suspension. The cells were counted manually under a microscope.
Cell Viability Assay
1×104 cells/well were seeded into a 96-well microplate, 18 h after seeding, treated with various concentrations (0, 0.5, 1, 2, and 4 μM) of pristimerin, triplicates per concentration. Twenty-four hours later, 10 μL of Cell Counting Kit-8 (CCK-8; Beyotimes, Wuhan, China) was added to each well. After incubation for 1 h, the optical density was tested by a spectrophotometer (Bio-Rad, Hercules, CA, USA) at 450 nm wavelength. The 0 μM pristimerin treatment was set as control cells, whereas medium alone was set as blank control. Cell viability was given by the following formula: Viability (% of control cells) = (A450 of treated cells - A450 of medium)/(A450 of control cells - A450 of medium)×100%.
Cell Apoptosis and Cell Cycle Detection by Flow Cytometry
Breast cancer cells were treated with 0 or 1 μM pristimerin for 24 h. Then, cell apoptosis and cell cycle were evaluated by Annexin V-FITC/PI apoptosis detection kit (BD Biosciences) and PI Staining Cell Cycle Detection Kit (Thermo Fisher, Waltham, MA, USA) following the manufacturer’s instructions,21 respectively. Finally, this was analyzed by using a FACS LSRFortessa flow cytometer. The 0 μM pristimerin treated group was set as control.
Cell apoptosis were analyzed with FlowJo 10.0 software package (Treestar Inc., Ashland, OR, USA). AnnexinV−/PI−, AnnexinV+/PI−, AnnexinV−/PI+, and AnnexinV+/PI+ were defined as live cells, early apoptosis, late apoptosis, and necrosis cells, respectively.22 DNA index (DI) as well as the estimation of cells in S and G2M cell cycle compartments were performed by automatic analysis using the ModFit LT 5.0.9 software (Verity Software House, USA).
Wound Healing Assay
Cells were seeded into 6-well plates and cultured with DMEM. After 12h, we scratched the plate quickly with a plastic tip, then changed the culture to serum-free DMEM with 0 or 1 μM pristimerin. Twenty-four hours later, wound closure was measured by ImageJ software. The 0 μM pristimerin treated group was set as control. Data was analyzed with ImageJ 1.52a software (National institutes of Health, USA). Also, we used a non-cytotoxic dose of 0.1 μM pristimerin treated MCF-7 cells for 5 h, then analyzed as mentioned above to determine the biological effect of pristimerin.
TIMER Analysis
The Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) algorithm is a database incorporating 10,009 samples across 23 cancer types from The Cancer Genome Atlas (https://cistrome.shinyapps.io/timer/).23 The gene module allows users to select any gene of interest and visualize the correlation of its expression in diverse cancer types. The survival module draws Kaplan-Meier plots for genes to visualize the survival differences. Levels are divided into low and high levels by a user-defined slider. Once all inputs are defined, TIMER outputs the Cox regression results including hazard ratios and statistical significance automatically. A P-value of Log rank test for comparing survival curves of two groups is shown. DUB3 expression in different cancer types was evaluated compared with normal tissues, also the relationship with DUB3 expression and overall survival (in breast cancer, or between breast cancer and normal) was analyzed.
Real-Time Polymerase Chain Reaction Analysis (RT-PCR)
First, total RNA of breast cancer cells was purified by using AxyPrep Multisource Total RNA Miniprep Kit (Axygen, Union City, CA, USA). Each sample was subjected to purity quantification using an OD 260/280 nm absorbance ratio. Pure samples with a 260/280 ratio between 1.8 and 2.1 were selected for subsequent experiments. Then, after checking the quality, RNA was converted to cDNA using PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Kusatsu, Japan). Polymerase chain reaction was performed using SYBR Green based on the instruction manual (Takara Bio, Kusatsu, Japan). Primer sequences were found in Primerbank (https://pga.mgh.harvard.edu/primerbank/), shown in Table 1 (Comate Bioscience, Changchun, China). Relative target mRNA expression was quantified using the ∆∆Ct method. U6 and β-actin were used as housekeeping genes.
 |
Table 1 Oligonucleotide and Primer Sequence |
Transfection with miR-542 Mimics
1×105 cells/well were seeded into 24-well microplate with DMEM. 18 h after seeding, 1 μL HiPerFect transfection reagent (Qiagen, Shanghai, China) with 2 μL 20 nM negative control or hsa-miR-542-5p mimics (GenePharma, Shanghai, China) were used for transfecting, mixed with 60 μL DMEM for 10 min, then medium was removed from the culture container, and the mixture above added to 150 μL medium with cells. Cells were harvested at 48 h after transfection. The transfection efficiency of hsa-miR-542-5p was confirmed by RT-PCR.
For cell viability detection, 1×104 cells/well were seeded into a 96-well microplate, 18 h after seeding, 0.3μL HiPerFect transfection reagent (Qiagen, Shanghai, China) with 0.6 μL 20 nM negative control or hsa-miR-542-5p mimics (GenePharma, Shanghai, China) were used for transfecting, mixed with 20 μL DMEM for 10 minutes, then removed medium from culture container, and the mixture above added to 100 μL medium with cells. After 24 h, cell activity was measured as described above, negative control was set as control.
Statistical Analysis
Statistical analyses were performed with Prism (version 8.0, GraphPad Software, San Diego CA, USA). Results were shown as mean±SD. Statistical comparisons were performed using two-way ANOVA or Student’s t-test of variance. P-values<0.05 were considered to denote statistical significance. All data represent at least three independent experiments.
Results
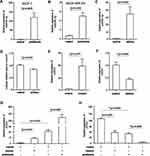
Pristimerin Inhibited the Growth of Breast Cancer Cells
Cell viability was performed to investigate the potential role of pristimerin by using CCK-8 assay (all P<0.05 with pristimerin concentrations from 1 μM up to 4 μM; Figure 1) in MCF-7, MDA-MB-231, and 4T1 breast cancer cells. The result showed that pristimerin did not significantly affect breast cancer cell viability at a concentration of 0–0.5 μM; however, cell proliferation was remarkably inhibited when the concentration was brought up to 1 μM. Whereas MCF-7 and MDA-MB-231 cell viability was obviously inhibited when 2 μM pristimerin was used in the experiment.
 |
Figure 1 Pristimerin inhibits the cellular viability. CCK-8 assay showed that pristimerin inhibited the viabilities of MCF-7, MDA-MB-231, and 4T1 breast cancer cells in a dose dependent manner. |
Pristimerin Inhibited Migration and Cell Cycle of Breast Cancer Cells, and Also Induced Apoptosis
Migration and metastasis are the majority cause of human cancer-related deaths.24 We used a wound healing assay to evaluate the migration ability of MCF-7 cells and found that under pristimerin treatment cell migration was markedly reduced in vitro (Figure 2A and B), suggesting a potential role of pristimerin in cell migration impeding. Furthermore, we used 0.1 μM pristimerin treated MCF-7 for 5 hours, and no significant changes were observed (Supplemental Figure S1), indicating the effect on cell migration is due to the cytotoxicity of pristimerin, other than the biological effect.
Pristimerin contributes to relaxation of the G1/S checkpoint upon decreasing cell cycle-related proteins expression in human cholangiocarcinoma cells.22 In line with this model, we used flow cytometric analysis of the cellular DNA content to discover the function of pristimerin on the cell cycle distribution and further elucidate the underlying mechanism of pristimerin in cell growth inhibition. We observed that MCF-7 cells exhibited G0/G1 phase of the cell cycle blocking under pristimerin treatment. In addition, S and G2‐M phase of the cell cycle also showed a concomitant reduction (Figure 2C and D). Overall, these results suggested that induction of G0/G1 phase arrest by pristimerin induced breast cancer cells growth inhibition.
To further evaluate the cell growth blocking function of pristimerin, we examined MCF-7 cells apoptosis by using Annexin V-FITC/PI double-staining and assessed fluorescence by flow cytometry. As shown in Figure 2E and F, pristimerin treatment significantly induced cell late apoptosis and necrosis, especially late apoptosis.
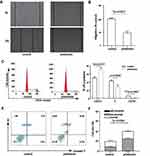
Pristimerin Down-Regulated DUB3 in Breast Cancer
In previous study, DUB3 inhibition was confirmed to suppresses breast cancer invasion and metastasis.25 Bioinformatic analysis by TIMER was used to evaluate DUB3 level in various cancer and normal tissues, as shown in Figure 3A. DUB3 was significantly overexpressed in breast cancer compared with normal. Next, the expression level of DUB3 in the overall survival analysis was calculated using the Kaplan-Meier curve. Compared to normal, breast cancer patients showed a higher expression of DUB3, with a lower overall survival rate (log-rank P=0.003), whereas higher expression of DUB3 showed a worse overall survival in breast invasion cancer, however with no significant difference (Figure 3B), indicating that DUB3 may play important roles in breast cancer and were related to survival rate. To investigate the potential role of pristimerin, the expression level of DUB3 with or without pristimerin treatment in MCF-7 and MDA-MB-231 were analyzed by RT-PCR. The results indicated that with anti-cancer effect, pristimerin treatment significantly decreased DUB3 level in both MCF-7 and MDA-MB-231 cells (Figure 3C and D). Our findings suggested that DUB3 is differently expressed in tumor and normal tissue of breast cancer and the level is associated with overall survival, under pristimerin treatment, DUB3 declined while cell malignancy decreased in breast cancer cells.
miR-542-5p Bounded to AGO2 and Targeted DUB3 in Breast Cancer Cells
The predominant function of miRNA has been confirmed to guide AGO2 to its specific targets through sequence complementarity, then located in RISC and lead to mRNA cleavage or translation blocking. miR-542 associated research was focused on proliferation, migration, metastasis, angiogenesis, and survival of tumor cells.26,28 Here, we treated MCF-7 and MDA-MB-231 human breast cancer cells with pristimerin, miR-542-5p expression was up-regulated (Figure 4A and B), while DUB3 was down-regulated. Hence, we hypothesized miR-542-5p might directly modulate AGO2 into RISC, subsequently the RISC leads to repression of DUB3, resulting in the proliferation, migration, and cell cycle were all inhibited by pristimerin. To verify the hypothesis, we used miR-542-5p transfection, then evaluated cell proliferation of breast cancer cells by CCK-8 assay. The result suggested that MCF-7 proliferation reduced with miR-542-5p overexpression (Figure 4C and FD). Next, we found AGO2 mRNA level was positive related with miR-542-5p, while DUB3 was down-regulated, suggesting that miR-542-5p might play upstream of AGO2 and DUB3 (Figure 4E and FF). Finally, the effect of pristimerin treatment and miR-542-5p overexpression in MCF-7 cells was evaluated. miR-542-5p mimics or control were transfected to MCF-7 cells, then treated with or without pristimerin (1 μM), RT-PCR was performed to determine AGO2 and DUB3 expression. 0 μM pristimerin treated with hsa-miR-542-5p control transfection group was set as negative control, whereas 1 μM pristimerin treated with hsa-miR-542-5p mimics transfection group was set as positive control. Our results revealed that overexpression miR-542-5p with pristimerin treatment elevate AGO2 and reduce DUB3 expression significantly (Figure 4G and H), therefore pristimerin and miR-542-5p overexpression had profoundly the same effect to inhibit breast cancer cells proliferation.
Discussion
Despite advances in diagnosis and treatment, breast cancer is still the most common cancer with high mortality in women, regardless of race or ethnicity.29 Pristimerin purified from traditional medicinal plants has the activity to inhibit tumor angiogenesis and prevent chemotherapy resistance.10,30 In the current study, we find pristimerin inhibits breast cancer cell proliferation, migration, and cell cycle, induces apoptosis, while DUB3 expression is down-regulated. Meanwhile breast cancer overexpression DUB3 compared with normal tissue, breast cancer patients with DUB3 high expression also exhibit a lower survival rate. In addition, we point out miR-542-5p is the key target in pristimerin reducing breast cancer cells viability, through DUB3 signal. miRNAs play an important role in the development of organisms and diseases, including both biological and cellular processes, such as malignant transformation. AGO2 is a crucial component of RISC and plays an essential role in RNA silencing process.31 RISC is known as RNA interference for gene-silencing ability. In some conditions, a large number of miRNAs expression levels could be affected under anti-tumor drugs treatment, shaping a specific miRNA expression profile footprint.32 As a novel anti-cancer pharmaceutical, pristimerin treatment may play a central regulatory role with differentially expressed miRNAs, eventually resulting in proliferation inhibited, G0/G1 phase arrest, and apoptotic cell death increased. Furthermore, these anti-cancer effects of pristimerin on breast cancer cells might alleviate or slow disease progression.
miR-542 was a multifunctional miRNA, which is involved in proliferation, invasion, cell apoptosis, and other biological processes.26,28 miR-542-5p and miR-542-3p are two mature sequences formed from pre-miR-542, while studies on miR-542-3p is abundant, studies on miR-542-5p is insufficient, whereas its role is still controversial. miR-542-3p is known as a tumor suppressor, overexpression of miR-542-3p may lead to proliferation, migration, angiogenesis, and survival inhibition, even decrease the metastatic potential of tumor cells.33,35 In the present study, we first discovered miR-542-5p expression was obviously higher under elevated pristimerin concentration accompanied with breast cancer cell viability reduced, which indicated that miR-542-5p might unfacilitate breast cancer cell proliferation. Subsequently, the miR-542-5p effect on RISC and proliferation associated gene DUB3 was evaluated. Our results revealed that pristimerin treatment and miR-542-5p overexpression shared synergistic effect, decreasing cell proliferation potentially achieved by its regulation on AGO2 and DUB3 expression, which was involved in breast cancer progression.
It is well documented that the dynamic equilibrium between ubiquitination and deubiquitination regulated most cellular protein stability, and ubiquitin-mediated protein degradation plays crucial roles in cancer-related cellular processes.36 As an important deubiquitinating enzyme, DUB3 is known to regulate a DNA damage response by controlling H2AX ubiquitination,37 stabilize Cdc25A protein phosphatase to couple the G1/S checkpoint,38 and stabilize SNAIL1 protein to induce epithelial–mesenchymal transition.39 Strikingly, it not only regulates CDK4/6 at post-translational level,39 but also combines to cyclin A to accelerate its deubiquitination and stabilize G1/S transition in cell cycle progression. Although the upstream of DUB3 is still unclear, the function of inhibiting cell proliferation and migration indicates that it may act as a novel therapeutic target in cancer. Here, we provided a mechanism basis for pristimerin application as an anticancer therapy, however, further studies are still needed.
Conclusions
In conclusion, the current study revealed the anti-cancer effect of pristimerin in breast cancer (participated in the proliferation, migration, cell cycle, apoptosis of cells) probably through miR-542-5p/DUB3 axis.
Acknowledgements
We appreciated critical comments and invaluable suggestions from Dr. Huanfa Yi's (Central Labloratory, The First Hospital of Jilin University) on this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Mu Z, Benali-Furet N, Uzan G, et al. Detection and characterization of circulating tumor associated cells in metastatic breast cancer. Int J Mol Sci. 2016;17(10):1665.
4. Sauter ER. Reliable biomarkers to identify new and recurrent cancer. Eur J Breast Health. 2017;13(4):162–167. doi:10.5152/ejbh.2017.3635
5. Brinker AM, Ma J, Lipsky PE, Raskin I. Medicinal chemistry and pharmacology of genus tripterygium (Celastraceae). Phytochemistry. 2007;68(6):732–766. doi:10.1016/j.phytochem.2006.11.029
6. Yousef BA, Hassan HM, Zhang LY, Jiang ZZ. Pristimerin exhibits in vitro and in vivo anticancer activities through inhibition of nuclear factor-small ka, CyrillicB signaling pathway in colorectal cancer cells. Phytomedicine. 2018;40:140–147. doi:10.1016/j.phymed.2018.01.008
7. Liu YB, Gao X, Deeb D, Arbab AS, Gautam SC. Pristimerin induces apoptosis in prostate cancer cells by down-regulating Bcl-2 through ROS-dependent ubiquitin-proteasomal degradation pathway. J Carcinog Mutagen. 2013;Suppl 6(3):005.
8. Wu CC, Chan ML, Chen WY, Tsai CY, Chang FR, Wu YC. Pristimerin induces caspase-dependent apoptosis in MDA-MB-231 cells via direct effects on mitochondria. Mol Cancer Ther. 2005;4(8):1277. doi:10.1158/1535-7163.MCT-05-0027
9. Tiedemann RE, Schmidt J, Keats JJ, et al. Identification of a potent natural triterpenoid inhibitor of proteosome chymotrypsin-like activity and NF-kappaB with antimyeloma activity in vitro and in vivo. Blood. 2009;113(17):4027–4037. doi:10.1182/blood-2008-09-179796
10. Xie G, Yu X, Liang H, et al. Pristimerin overcomes adriamycin resistance in breast cancer cells through suppressing Akt signaling. Oncol Lett. 2016;11(5):3111–3116. doi:10.3892/ol.2016.4335
11. Yousef BA, Hassan HM, Zhang LY, Jiang ZZ. Anticancer potential and molecular targets of pristimerin: a mini- review. Curr Cancer Drug Targets. 2017;17(2):100–108. doi:10.2174/1568009616666160112105824
12. Paulmurugan R, Oronsky B, Brouse CF, Reid T, Knox S, Scicinski J. Real time dynamic imaging and current targeted therapies in the war on cancer: a new paradigm. Theranostics. 2013;3(6):437–447. doi:10.7150/thno.5658
13. Trujillo RD, Yue S-B, Tang Y, O’Gorman WE, Chen C-Z. The potential functions of primary microRNAs in target recognition and repression. EMBO J. 2010;29(19):3272–3285. doi:10.1038/emboj.2010.208
14. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
15. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi:10.1016/j.molmed.2014.06.005
16. Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78(1):363–397. doi:10.1146/annurev.biochem.78.082307.091526
17. Pereg Y, Liu BY, O’Rourke KM. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol. 2010;12(4):400–406. doi:10.1038/ncb2041
18. Zhang Q, Zhang ZY, Du H, et al. DUB3 deubiquitinates and stabilizes NRF2 in chemotherapy resistance of colorectal cancer. Cell Death Differ. 2019;26(11):2300–2313.
19. Wu X, Luo Q, Zhao P. MGMT-activated DUB3 stabilizes MCL1 and drives chemoresistance in ovarian cancer. Proc Natl Acad Sci U S A. 2019;116(8):2961–2966. doi:10.1073/pnas.1814742116
20. Hu B, Deng T, Ma H, et al. Deubiquitinase DUB3 regulates cell cycle progression via stabilizing cyclin A for proliferation of non-small cell lung cancer cells. Cells. 2019;8(4):297. doi:10.3390/cells8020137
21. Lin KH, Xie A, Rutter JC, et al. Systematic dissection of the metabolic-apoptotic interface in AML reveals heme biosynthesis to be a regulator of drug sensitivity. Cell Metab. 2019;29(5):1217–1231.
22. Sun JM, Xu HT, Zhao L, et al.Induction of cell-cycle arrest and apoptosis in human cholangiocarcinoma cells by pristimerin. J Cell Biochem. 2019;120(7):12002–12009.
23. Li B, Severson E, Pignon J-C. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
24. Chaffer C, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi:10.1126/science.1203543
25. Wu Y, Wang Y, Lin Y. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat Commun. 2017;8(1):14228. doi:10.1038/ncomms14228
26. Rivero RM, Kojima M, Gepstein A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci. 2007;104(49):19631–19636. doi:10.1073/pnas.0706963104
27. Schmeier S, MacPherson CR, Essack M, et al. Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genomics. 2009;10(1):595. doi:10.1186/1471-2164-10-595
28. Singh R, Kaushik S, Wang Y. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi:10.1038/nature07976
29. Fares J, Kanojia D, Rashidi A, et al. Diagnostic clinical trials in breast cancer brain metastases: barriers and innovations. Clin Breast Cancer. 2019;19(6):383–391.
30. Mu X, Shi W, Sun L, Li H, Jiang Z, Zhang L. Pristimerin, a triterpenoid, inhibits tumor angiogenesis by targeting VEGFR2 activation. Molecules. 2012;17(6):6854. doi:10.3390/molecules17066854
31. Jang JH, Jung JS, Im YB, Kang KS, Choi JI, Kang SK. Crucial role of nuclear Ago2 for hUCB-MSCs differentiation and self-renewal via stemness control. Antioxid Redox Signal. 2012;16(2):95–111. doi:10.1089/ars.2011.3975
32. Gillet V, Hunting DJ, Takser L. Turing revisited: decoding the microRNA messages in brain extracellular vesicles for early detection of neurodevelopmental disorders. Curr Environ Health Rep. 2016;3(3):188–201. doi:10.1007/s40572-016-0093-0
33. Althoff K, Lindner S, Odersky A, et al. miR-542-3p exerts tumor suppressive functions in neuroblastoma by downregulating Survivin. Int J Cancer. 2015;136(6):1308–1320. doi:10.1002/ijc.29091
34. Zhang J, Wang S, Han F, et al. MicroRNA-542-3p suppresses cellular proliferation of bladder cancer cells through post-transcriptionally regulating survivin. Gene. 2016;579(2):146–152. doi:10.1016/j.gene.2015.12.048
35. Wang Y, Huang JW, Castella M, Huntsman DG, Taniguchi T. p53 is positively regulated by miR-542-3p. Cancer Res. 2014;74(12):3218–3227. doi:10.1158/0008-5472.CAN-13-1706
36. Hu B, Deng T, Ma H, et al. Deubiquitinase DUB3 regulates cell cycle progression via stabilizing cyclin A for proliferation of non-small cell lung cancer cells. Cells. 2019;8(4):297. doi:10.3390/cells8040297
37. Delgado-Díaz MR, Martín Y, Berg A, Freire R, Smits VAJ. Dub3 controls DNA damage signalling by direct deubiquitination of H2AX. Mol Oncol. 2014;8(5):884–893. doi:10.1016/j.molonc.2014.03.003
38. van der Laan S, Tsanov N, Crozet C, Maiorano D. High Dub3 expression in mouse ESCs couples the G1/S checkpoint to pluripotency. Mol Cell. 2013;52(3):366–379. doi:10.1016/j.molcel.2013.10.003
39. Liu T, Yu J, Deng M, et al. CDK4/6-dependent activation of DUB3 regulates cancer metastasis through SNAIL1. Nat Commun. 2017;8(1):13923. doi:10.1038/ncomms13923
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.