Back to Journals » International Journal of Nanomedicine » Volume 15
Primary Amine Modified Gold Nanodots Regulate Macrophage Function and Antioxidant Response: Potential Therapeutics Targeting of Nrf2
Authors Luo YH , Cheng HJ, Tsai FY, Tsou TC, Lin SY , Lin P
Received 17 June 2020
Accepted for publication 2 October 2020
Published 29 October 2020 Volume 2020:15 Pages 8411—8426
DOI https://doi.org/10.2147/IJN.S268203
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Yueh-Hsia Luo,1 Hsien-Jen Cheng,2 Feng-Yuan Tsai,2 Tsui-Chun Tsou,2 Shu-Yi Lin,3 Pinpin Lin2
1Department of Life Sciences, National Central University, Taoyuan City 32001, Taiwan; 2National Institute of Environmental Health Sciences, National Health Research Institutes, Zhunan, Miaoli County 35053, Taiwan; 3Institute of Biomedical Engineering and Nanomedicine, National Health Research Institutes, Zhunan, Miaoli County 35053, Taiwan
Correspondence: Yueh-Hsia Luo
Department of Life Sciences, National Central University, No. 300, Zhongda Road, Zhongli District, Taoyuan City 32001, Taiwan
Tel +886-34227151
Fax +886-34228482
Email [email protected]
Pinpin Lin
National Institute of Environmental Health Sciences, National Health Research Institutes, 35 Keyan Road, Zhunan, Miaoli County 35053, Taiwan
Tel +886-37246166
Fax +886-37587406
Email [email protected]
Background: Gold nanoparticles with high biocompatibility and immunomodulatory properties have potential applications in the development of new diagnostic and therapeutic strategies for nanomedicine. Nanoparticles targeting macrophages can manipulate or control immunological diseases. This study assessed the activity of dendrimer-encapsulated gold nanodots (AuNDs) with three surface modifications [ie, outfacing groups with primary amine (AuNDs-NH2), hydroxyl (AuNDs-OH), and quaternary ammonium ions (AuNDs-CH3)] regulated macrophage function and antioxidant response through Nrf2-dependent pathway.
Methods: AuNDs were prepared and characterized. Intracellular distribution of AuNDs in human macrophages was observed through confocal microscopy. The activity of AuNDs was evaluated using macrophage functions and antioxidant response in the human macrophage cell line THP-1.
Results: AuNDs-NH2 and AuNDs-CH3, but not AuNDs-OH, drove the obvious Nrf2-antioxidant response element pathway in THP-1 cells. Of the three, AuNDs-NH2 considerably increased mRNA levels and antioxidant activities of heme oxygenase 1 and NAD(P)H quinone dehydrogenase 1 in THP-1 cells. IL-6 mRNA and protein expression was mediated through Nrf2 activation in AuNDs-NH2-treated macrophages. Furthermore, Nrf2 activation by AuNDs-NH2 increased the phagocytic ability of THP-1 macrophages.
Conclusion: AuNDs-NH2 had immunomodulatory activities in macrophages. The findings of the present work suggested that AuNDs have potential effects against chronic inflammatory diseases via the Nrf2 pathway.
Keywords: gold nanodots, macrophage, antioxidant, inflammation
Introduction
Engineering nanoparticles for application in the immune system is an exciting, emerging field in the development of nanotechnology.1 Nanoparticles targeting immune cells (eg, macrophages) can help in manipulating or controlling immunological diseases, such as infectious diseases or tumors. Macrophages play many key roles in the host defense system. They not only phagocytose dead cells and pathogens but also shape the inflammatory response and modulate adaptive immunity. However, macrophages act as major effector cells in chronic inflammatory development. Redox signals affect the process of macrophage polarization and reprogramming.2 A key master redox regulator is the nuclear factor erythroid 2-related factor 2 (Nrf2) signaling pathway, responsible for oxidative and xenobiotic stress.3 Moreover, Nrf2 is a vital target for anti-inflammation and antioxidant cytoprotective pathways.4,5 Nrf2 activators exhibit protective effects in animal models of neurodegenerative diseases6–8 and chronic obstructive pulmonary disease.9 Accordingly, natural or synthetically derived Nrf2 modulators have been attracting attention as a promising therapeutic strategy for chronic inflammatory diseases.10
The extent of biocompatibility of a nanoparticle depends on its various surface properties. Owing to the inert and nontoxic core, gold nanoparticles usually exhibit relatively low toxicity and high biocompatibility compared with other metal-based nanoparticles, such as silver, zinc oxide, and iron oxide.11,12 Our previous study demonstrated that surface modification of gold nanodots (AuNDs) changed the lipopolysaccharide (LPS) assembly behavior to modulate immunological effects in human macrophages.13 Liu et al14 also demonstrated that surface modifications of AuNDs with amine outfacing group (AuNDs-NH2) demonstrated reduced peroxidase-like activity while still retaining catalase-like activity. The methylation of AuNDs-NH2 to form quaternary ammonium ions (AuNDs-CH3) restored the intrinsic peroxidase-like activity.
In the present study, AuNDs with three types of outfacing groups—namely primary amine (AuNDs-NH2), hydroxyl (AuNDs-OH), and quaternary ammonium ions (AuNDs-CH3)—had different effects antioxidant response and macrophage functions via the Nrf2 activation pathway. Of the three, only AuNDs-NH2 triggered transient reactive oxygen species (ROS) disturbance within 10 min and then decreased intracellular ROS in a time-dependent manner. AuNDs-NH2 induced Nrf2 ser40 phosphorylation and subsequently triggered Nrf2 target gene transcriptional activation, and AuNDs-CH3 activated Nrf2 through the noncanonical Nrf2-P62 pathway. However, AuNDs-OH did not trigger Nrf2- antioxidant response element (ARE) activation. AuNDs-NH2 considerably increased hemeoxygenase 1 (HO-1) and NAD(P)H quinone dehydrogenase 1 (NQO1) mRNA levels in THP-1 cells after 24 h of treatment. In addition, the antioxidant enzyme activities were elevated in AuNDs-NH2-treated cells at 48 h, including NQO1, glutathione peroxidase (GPx), glutathione S-transferase (GST), and catalase. Only NQO1 and catalase enzyme activities were increased after AuNDs-CH3 treatment. Moreover, mRNA and protein expression of IL-6 was mediated through Nrf2 activation in AuNDs-NH2-treated macrophages. Furthermore, Nrf2 activation by AuNDs-NH2 increased the phagocytic ability of THP-1 macrophages. Therefore, our findings provide rationale strategies for the development of nanomaterials targeting the Nrf2 pathway against chronic diseases.
Materials and Methods
Materials
G4NH2 and G4OH dendrimers, HAuCl4, methyl iodide, LPS, and ML385 were purchased from Sigma-Aldrich (St. Louis, MO, USA). 2ʹ,7ʹ dichlorodihydrofluorescein diacetate (DCFH-DA) was obtained from Thermo Fisher Scientific (Grand Island, NY, USA). The AuNDs were synthesized according to a previous study.15 In brief, G4NH2 and HAuCl4 were mixed, and the solution was incubated at 4 °C overnight and then irradiated with microwave at 120 °C for 30 min (CEM; Discover LabMate System). The precipitations and AuNDs were filtered through an MWCO membrane filter (3 KDa) from Millipore (PES membrane, MA, USA). The extra AuCl4− was removed using anionic exchange chromatography (Fractogel EMD TMAE Hicap; Rockland, Merck, MA, USA). The internal tertiary amine groups and the surface amine groups of dendrimer-encapsulated AuNDs were treated with methyl iodide in 1 mL of 1:1 (v/v) N,N′-dimethylformamide/H2O at room temperature overnight to form AuNDs-CH3. The reaction mixture was extracted, dried under a vacuum, and dissolved in water.14
Physicochemical Properties of AuNDs
Zeta potential was measured on the Zetasizer Nano system (Zetasizer Nano ZS; Malvern Instruments, Worcestershire, UK). All measurements were performed with three independent experiments at room temperature. The gold content of AuNDs and percentages of cell uptake were quantified through inductively coupled plasma mass spectrometry (ICP-MS). Polarities of AuNDs were measured using pyrene, a type polarity probe.16–18 The I1:I3 ratio is an index of the polarity of its environment; I1 and I3 are intensities at 372 and 383 nm, respectively. AuND sizes were determined on a dynamic light scattering instrument (Malvern Zetasizer, Nano ZS) with an argon laser wavelength (λ = 633 nm; detector angle = 173°; typical sample volume = 100 μL). Endotoxin levels of AuNDs were measured using the kinetic turbidimetric assay. The spike recoveries of AuNDs-NH2, AuNDs-CH3, and AuNDs-OH were 86%, 85%, and 93%, respectively.
Intracellular Distribution of AuNDs
THP-1 cells were purchased from Bioresource Collection and Research Centre in Food Industry Research and Development Institute, Taiwan. The cells were seeded in a 3.5-cm μ-dish (ibidi GmbH, Martinsried, Germany) and treated for 3 days with 100 nM PMA. After replacement with fresh serum-free medium for 1 day, the cells were treated with 1 mg/mL AuNDs for 6 and 24 h. MitoTracker (Invitrogen, Molecular Probe, OR, USA) and LysoTracker (Invitrogen) were used for intracellular biodistribution of AuNDs. MitoTracker is a cell-permanent probe that contains a mildly thiol-reactive chloromethyl moiety for labeling mitochondria. LysoTracker is a fluorescent acidotropic probe for labeling and tracking acidic organelles in live cells and can be used to investigate intracellular lysosomes. The red MitoTracker and LysoTracker were detected with confocal microscopy upon excitation using a 577-nm diode laser, and the emission was measured at 590 nm. The intracellular distributions of the AuNDs were detected with confocal microscopy on excitation using a 405-nm diode laser, and the emissions were measured at 420–460 nm. To examine whether AuNDs are excreted through exocytosis or destroyed in cells, PMA-activated THP-1 cells were treated with 1 mg/mL AuNDs for 24 h. Then, AuNDs were removed by replacement with a new culture medium for another 48 h. The cells were stained with LysoTracker (red) and then examined using confocal microscopy.
Measurement of Intracellular ROS
ROS generation was determined using DCFH-DA. PMA-activated THP-1 cells were plated with an appropriate number in 6-well plates or 96-well black microtiter plates and treated for 10 and 30 min with or without AuNDs or LPS, diluted directly into the culture media. Treatment with 1 μg/mL LPS for 30 min was used as a positive control. An appropriate number of cells were incubated with 5 μM DCFH-DA at 37 °C for 30 min and then immediately measured using a microplate reader or through flow cytometry with a 485-nm excitation wavelength and 520-nm emission wavelength.
Adenovirus-Based ARE Luciferase Reporter Assay
The use of adenovirus-based luciferase reporter assay has been established.19,20 In brief, the PMA-activated cells (1.5 × 104 cells/well) were seeded on 96-well white plates (cat. no. 136101; Nalge Nunc, Roskilde, Denmark). After infection with the recombinant adenovirus containing ARE constructs at a multiplicity of infection of 1 in serum-free medium for 16 h, the infected THP−1 cells were treated with AuNDs for 24 h. Luciferase activity was determined in triplicate on the Luciferase Assay System (Promega, Madison, WI, USA) by using a programmed microplate luminometer21 (MicroLumatPlus LB96V, EG&G Berthold, Germany).
Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction Assays
THP−1 cells were seeded in 6-well plates and then treated for 3 days with 100 nM PMA. After replacement with fresh sera-free medium for 1 day, the cells were treated with 1 mg/mL AuNDs for 24 or 48 h. Next, RNA was extracted using an RNAzol RT kit (Life Technologies, Rockville, MD, USA). The purified RNAs were stored at −80 °C, and cDNA was synthesized with total RNA (3 μg). Quantitative real-time reverse transcription-polymerase chain reaction (quantitative PCR) was used to measure NQO1, HO−1, IL−1β, and IL−6 expression, and the assays were performed using the Assay-on-Demand Gene Expression Assay Mix (Applied Biosystems, Foster City, CA, USA). The assay ID for NQO1 was Hs00168547_m1, of HO−1 was Hs00157965_m1, for IL−1β was Hs01555410_m1, for IL−6 was Hs00174131_m1, and for glyceraldehyde−3-phosphate dehydrogenase (GAPDH) was Mm99999915_g1. Quantitative PCR to measure NQO1, HO−1, IL−1β, IL−6, and GAPDH expression were performed using TaqMan Universal PCR Master Mix (Applied Biosystems). The reaction mixtures were prepared by mixing aliquots of cDNA, 0.5 μL of Assay-on-Demand Gene Expression Assay Mix, and 5 μL of TaqMan Universal PCR Master Mix (Applied Biosystems) in a final volume of 10 μL. The reaction mixtures were analyzed on an ABI PRISM 7900 Sequence Detector System (Applied Biosystems) with the following PCR program: 95 °C for 10 min, followed by 40 cycles of 60 °C for 1 min, and 95 °C for 15 s. Quantitative values were obtained from the threshold cycle (Ct) number. The relative mRNA levels of the target genes were derived using the eq 2−ΔCt, where ΔCt = Cttarget gene − CtGAPDH. Data were presented as the fold relative to the control value.22
IL-1β and IL-6 ELISA
THP-1 cells were seeded in 6-well plates and then treated for 3 days with 100 nM PMA. After replacement with fresh sera-free medium for 1 day, the cells were treated with 1 mg/mL AuNDs for 48 h. Human IL-1β and IL-6 proteins were measured using ELISA (Affymetrix eBioscience), according to manufacturer instructions.
Antioxidant Enzyme Activity Assays
Antioxidant enzyme activities were determined in AuNDs-treated cell at 48 h. The measured enzymes included NAD(P)H dehydrogenase [quinone] 1 (NQO1), glutathione peroxidase (GPx), catalase, and glutathione-S-transferase (GST). The enzyme activities of NQO1, GPx, catalase, and GST were determined using enzyme activity assays (Abcam, Cambridge, MA, USA), according to the instruction manuals.
Phagocytosis Assay
The phagocytosis assay was performed with pHrodo Escherichia coli BioParticles conjugates (Thermo Fisher Scientific). THP-1 cells were activated by PMA for 3 days and cultured with sera-free medium 24 h after PMA removal. THP-1 cells were treated with AuNDs for 24 or 48 h and then harvested from the culture plates for the following assay. The cells were incubated with pHrodo E. coli (1 mg/mL) at 37 °C for 2 h. The pHrodo dye conjugates were nonfluorescent outside the cell but brightly red fluorescent in phagosomes. The percentage of phagocytosis was detected through flow cytometry with the wavelength of excitation at 560 nm and emission at 585 nm.
Nrf2 and P62 Phosphorylations Through Flow Cytometry
Nrf2 ser40 and P62 ser349 phosphorylations were determined through flow cytometry. THP-1 cells were activated using PMA for 3 days and cultured with sera-free medium for another 24 h after PMA removal. THP-1 cells were treated with AuNDs at various time points. The cells were fixed with a paraformaldehyde buffer followed through methanol permeabilization. Anti-Nrf2 (phospho S40) and anti-SQSTM1/p62 (phospho S349) antibodies were purchased from Abcam. Anti-rabbit IgG conjugated with Alexa Fluor 488 (Invitrogen) was used as the secondary antibody. The percentage of Nrf2 and P62 phosphorylation was detected through flow cytometry with excitation wavelength of 495 nm and emission wavelength of 519 nm.
Statistical Analysis
All statistical analyses were conducted on Prism 4.0 (GraphPad Software, San Diego, CA, USA). Significant differences between treatment groups were determined through using an unpaired t-test. The results of experimentally treated groups and their corresponding controls were compared using one-way analysis of variance. Differences were considered statistically significant at *P values of <0.05, **P <0.01, and ***P <0.001.
Results
Physiochemical Properties and Intracellular Distribution of AuNDs in THP-1 Macrophages
Our previous study indicated that dendrimer-encapsulated AuNDs are highly biocompatible with macrophages.13 The preparations of AuNDs are described in the previous report,15 in which the detailed physicochemical properties are examined. In brief, the dimensions of AuNDs are uniformly smaller than the parent dendrimers (ie, G4NH2 and G4OH). Both AuNDs-NH2 and AuNDs-CH3 diminish the intrinsic cytotoxicity of G4NH2 through an irreversible backfolding of outfacing 1°-amines. Such a significant deactivation of the external 1°-amines can minimize the interaction between G4NH2 and cell membranes during internalization. Because of the absence of 1°-amines within the G4OH structure, AuNDs-OH are also highly biocompatible.15 Our previous reports examined the morphology and photoluminescence of AuNDs.23–25 The photoluminescence of AuNDs peaks at approximately 460 nm in an aqueous solution.26 The size of a single AuND is theoretically <1 nm, making it difficult to observe particle size using high-resolution transmission electron microscopy. Yeh et al23 found that AuNDs form a layer-by-layer film on the transmission electron microscopy copper grid. The proximity distance range of the gold atoms was speculated to be nearly 0.2884 nm.27 Other physicochemical properties, endotoxin levels, and uptake percentage of the three AuNDs are summarized in Table 1.
 |
Table 1 Physicochemical Properties, Endotoxin Levels, and Uptake Percentage of AuNDs |
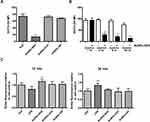
In summary, all three AuNDs exhibited positive zeta potential. The gold content of AuNDs-NH2 and AuNDs-CH3 was higher than that of AuNDs-OH. Polarity assays were performed using a well-known polarity probe (ie, pyrene)28 to measure the hydrophobicity of AuNDs. The polarity change from the microenvironment was indicated by two vibrational peaks of pyrene appearing at approximately 370 nm (I1) and 380 nm (I3). Our results revealed that AuNDs-OH is more hydrophobic than AuNDs-NH2 and AuNDs-CH3. Because our cell model is a macrophage, we also quantified the endotoxin levels of the three AuNDs to ensure that our experiments were free of LPS contaminants. The endotoxin levels were <0.005 (EU/mL) in AuNDs-NH2, 0.00825 (EU/mL) in AuNDs-CH3, and 0.01826 (EU/mL) in AuNDs-OH. The endotoxin levels of all three AuNDs were lower than the clinically acceptable concentration (0.25 EU/mL). Furthermore, the percentages of cellular uptake after AuND treatments were determined through ICP-MS. After 24 h of treatment, the cellular uptake percentage of AuNDs-OH was the lowest among the three AuNDs.
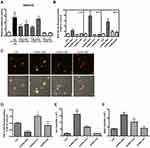
Next, we investigated the intracellular distributions in THP-1 macrophages after 24 h of treatment. All three AuNDs accumulated in the intracellular lysosome (Figure 1A) after entering macrophages and did not obviously affect or accumulate in the mitochondria (Figure 1B). No AuNDs influenced mitochondrial ATP production after 24 h of treatment (data not shown). We assessed whether AuNDs are retained in the cells or excreted through exocytosis. THP-1 cells were treated with AuNDs for 24 h, and then AuNDs were removed by replacement with a new culture medium for another 48 h. The results revealed that all AuNDs were retained in lysosomes after 72 h of treatment (Figure 1C).
AuNDs-NH2 Reduce Intracellular ROS Levels in Macrophages
The cellular redox system is involved in various physiological homeostatic mechanisms, such as cell differentiation, activation, death, and survival.29,30 Major endogenous ROS generation occurs in various organelles, such as the mitochondrial electron transport chain, endoplasmic reticulum system, and NADPH oxidase complex.30 Excessive ROS production triggers oxidative stress, which may result in lipid peroxidation, DNA damage, and protein dysfunction, leading to various disorders.31 Several studies have reported that nanoparticles can increase intracellular ROS levels and induce oxidative stress.32 We determined the intercellular ROS levels after AuNDs’ entry into macrophages. Our results revealed that ROS levels decreased in cells treated by AuNDs-NH2, but not AuNDs-CH3 or AuNDs-OH, after 24 h of treatment (Figure 2A) and in a time-dependent manner (Figure 2B). Furthermore, we examined the change in ROS levels in THP-1 cells within 30 min of treatment with AuNDs-NH2 and found that intracellular ROS levels transiently increased at 10 min but returned to those of the control group at 30 min (Figure 2C). LPS treatment was a positive control for ROS production within 30 min in macrophages. These results indicate that AuNDs-NH2 cause short-term ROS disturbance and then significantly reduce the intracellular ROS content. Therefore, we speculate that AuNDs-NH2 may regulate antioxidant mechanisms in macrophages.
Nrf2 Activation and Phosphorylation Occur in AuNDs-NH2-Treated Cells
Nrf2 is a master regulator of antioxidant responses. Nrf2 localizes within the cytoplasm and interacts with Kelch-like ECH-associating protein 1 (Keap1), which subsequently is ubiquitinated and degraded in the proteasome under normal conditions. In the canonical Keap1/Nrf2 pathway, oxidative stress or electrophile compounds modify Keap1 cysteine residues, leading to Keap1 conformation changes. Next, Nrf2 translocates to the nucleus and binds to the ARE or electrophile responsive element (EpRE), leading to target gene expression.3,30 In the noncanonical Nrf2-ARE activation pathway, a regulator of autophagy/xenophagy, p62/SQSTM1, facilitates Keap1 degradation and subsequently stabilizes and translocates Nrf2 into the nucleus.28 Nrf2 expression can be upregulated through Keap1-independent mechanisms, which include other transcriptional (eg, AHr-ARNT and NF-κB),33,34 posttranscriptional (microRNAs),35–38 and posttranslational (eg, phosphorylation and acetylation) modifications.39,40 We used the ARE reporter system to evaluate whether AuNDs activate the Nrf2 pathway in THP-1 cells and found that both AuNDs-NH2 and AuNDs-CH3 can activate Nrf2 (Figure 3A). Because none of the three types of AuNDs caused oxidative stress in cells and AuNDs-NH2 only transiently triggered ROS disturbance at 10 min, we speculated that AuNDs may activate Nrf2 through Keap1-independent mechanisms by activating intracellular kinases or other noncanonical pathways, such as the autophagic pathway–associated protein P62. Our results revealed that AuNDs-NH2 caused Nrf2 Ser40 phosphorylation, and Nrf2 translocated to the nucleus (Figure 3B and C) through the canonical pathway.
Ser349 phosphorylation of P62 binds to Keap1, separating Nrf2 from Keap1, and Nrf2 can then enter the nucleus to activate downstream genes through noncanonical pathways. Blockage of autophagosome maturation and accumulation of P62 result in prolonged Nrf2 activation in a P62-concentration-dependent manner.41 We found that AuNDs-CH3 induced P62 Ser349 phosphorylation (Figure 3D), suggesting that AuNDs-CH3 activate Nrf2 through the P62–Keap1–Nrf2 noncanonical pathway. P62 is an autophagy adaptor protein, which mediates the formation of protein aggregates intended for autophagy turnover. This means that P62 facilitates selective degradation of protein cargo through activating autophagy.41 The direct interaction between P62 and Keap1 allows Keap1 sequestration into the autophagosomes, leading to impairment of Nrf2 ubiquitylation and activation of the Nrf2 signaling pathway.41 Our result indicated that AuNDs-NH2 and AuNDs-CH3 may activate Nrf2 through different pathways. However, the role of Nrf2 activation by AuNDs-CH3 through the P62-dependent pathway needs further study. Moreover, we found that AuNDs-NH2 and AuNDs-CH3 can induce mRNA levels of HO-1 and NQO1 in THP-1 cells, but the increase in the levels of HO-1 and NQO1 in AuNDs-NH2-treated cells was significantly greater than that in AuNDs-CH3-treated cells (Figure 3E and F).
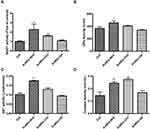
AuNDs-NH2 Activate Antioxidant Enzyme Activities
We further examined the activities of several Nrf2-mediated antioxidant enzymes in AuNDs-treated cells. Our results revealed that the activities of NQO1, GPx, catalase, and GST increased after 48 h of AuNDs-NH2 treatment (Figure 4). Similarly, AuNDs-CH3 increased activities of NQO1 and catalase (Figure 4A and D). Next, we used a Nrf2 inhibitor, ML385, to investigate whether the Nrf2 activations can be restored in AuNDs-NH2-treated cells. ML385 is mainly bound with the Neh1 domain of Nrf2, which interferes with the binding of Nrf2 and thus affects the combination of Nrf2 and the ARE element.42 Our results revealed that mRNA levels of HO-1 and NQO1 were reduced through pretreatment with the ML385 inhibitor (Figure 5B and C). tBHQ was a positive control for Nrf2 activation (Figure 5A). However, ML385 did not inhibit HO-1 mRNA expression in AuNDs-CH3-treated cells (Figure 5B). This indicates that AuNDs-CH3 may also affect other pathways involved in the regulation of HO-1. In addition to Nrf2, several transcription factors such as activator protein-1, nuclear factor-κB, and some upstream kinases have been identified as regulators of HO-1 gene induction.43
Nrf2 Modulated Expression of Proinflammatory Cytokines in AuNDs-Treated Macrophages
Many studies have suggested that Nrf2 is essential for the control of inflammation.4,5,44,45 Kobayashi et al5 reported that Nrf2 binds to the upstream regions of the IL6 and IL1b genes leading to interferes with LPS-induced transcriptional upregulation of proinflammatory cytokines. Therefore, we investigated the expression levels of these proinflammatory genes in AuNDs-treated cells. We found that AuNDs-NH2 upregulated IL-1β and IL-6 mRNA and protein expression (Figure 6). The Nrf2 inhibitor ML385 significantly reduced the AuNDs-NH2-induced increase in IL-6 expression in macrophages (Figure 6B and D). However, ML385 pretreatment did not affect IL-1β expression in AuNDs-NH2-treated cells (Figure 6A and C). ML385 blocked Nrf2 transcriptional activity by binding to the DNA-binding domain of Nrf2. AuNDs-NH2 increased the expression of IL-1β from 50 to 150 pg/mL and of IL-6 from 10 to 25 pg/mL. Nrf2 is required for the assembly of NLRP3 inflammasome activators, including the apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) adaptor, which may promote IL-1β production.46,47 Our data indicated that AuNDs-NH2-induced increased IL-1β productions may be irrelevant to the transcriptional control of Nrf2. It was reported that the ARE element sequence is found in the promoter region of IL-6, so Nrf2 can regulate IL-6 expression.48 Our result indicated that AuNDs-NH2 directly regulates IL-6 expression via a Nrf2-dependent pathway.
AuNDs-NH2 Increase Phagocytic Ability in Macrophages
In addition to the regulation of antioxidant and inflammatory responses, Nrf2 was found to regulate the phagocytic capacity of immune cells.49 Our result indicated that AuNDs-NH2 increased the phagocytic ability of human macrophages after 24 and 48 h of treatment. AuNDs-CH3 and AuNDs-OH also slightly affected phagocytosis at 24 h, but their effect was lower even than that of the control group at 48 h after treatment (Figure 7). ML385 reduced the effect of AuNDs-NH2 on phagocytosis, suggesting that Nrf2 participates in macrophage phagocytosis.
Discussion
Our results (Figure 8) revealed that AuNDs mainly accumulate in lysosomes. Nanoparticle-induced oxidative stress responses are torchbearers for further pathophysiological effects, including genotoxicity, inflammation, and fibrosis, through activation of associated cell signaling pathways.50 None of the three AuNDs induced significant oxidative stress in macrophages. Only AuNDs-NH2 transiently increased ROS levels at 10 min after treatment and then decreased intracellular ROS levels in a time-dependent manner. We speculated that this transient ROS perturbation may activate intracellular kinases to promote phosphorylation of Nrf2 Ser40, followed by the activation of Nrf2 downstream genes and the increase in antioxidant and phagocytic abilities.
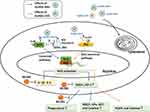 |
Figure 8 Pictorial description of how the AuNDs mediated Nrf2, antioxidant enzyme activities, and phagocytosis abilities in macrophages. |
Nrf2 plays a vital role in regulating cellular antioxidant and other cytoprotective responses, including purine, pentose, and lipid metabolisms, inflammation, and proteostasis,51 as well as cytoprotective mechanisms against various environmental stresses.52 Nrf2 is sequestered in the cytosol by Keap1 and constantly degraded by the ubiquitin–proteasome system under normal physiological conditions.53 Oxidative stress and chronic inflammation may be as primary etiologic factors in many chronic diseases.54 Nrf2 modulators have been attracting attention as a promising therapeutic strategy for chronic diseases dominated by oxidative stress and inflammation, such as neurodegenerative,55 cardiovascular, and metabolic diseases.10,51,56 Activation of endogenous antioxidant gene expression, modulated via the Nrf2 pathway, has a better neuroprotective effect than that afforded by conventional antioxidant therapy.57–59 In experimental models of asthma and chronic obstructive pulmonary disease (COPD), Nrf2 targeting also suppresses airway inflammation and oxidative stress.60,61
In macrophage, Nrf2 acts as the upstream regulator of inflammatory cytokine production in a ROS-independent manner.5 Moreover, Nrf2 directly regulates the expression of several other macrophage-specific genes that are associated with phagocytosis and scavenger receptor for oxidized low-density lipoprotein.49,62 Our results indicated that AuNDs-NH2 upregulated IL-1β and IL-6 mRNA and protein expression. Because the ARE element sequence is found in the promoter region of IL-6, Nrf2 may regulate IL-6 expression.51 ML385, an inhibitor of Nrf2 transcriptional activity, significantly reduced AuNDs-NH2-mediated Nrf2 activation and expression of NQO1, HO-1, and IL-6 mRNA, as well as the phagocytic ability in macrophages. AuNDs-NH2-induced increased IL-1β production may not occur through Nrf2 transcriptional regulation. However, further research is needed to determine whether AuNDs-NH2 activate Nrf2 to prevent LPS-induced proinflammatory cytokines. In addition, only AuNDs-NH2 increase phagocytic ability in macrophages via a Nrf2-mediated pathway. Targeting Nrf2 signaling improves infection clearance by enhancing phagocytosis activity, which might be a therapeutic target. Harvey et al demonstrated that sulforaphane, which targets Nrf2 signaling, improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model.49 Furthermore, in a mouse model of papain-induced rhinosinusitis, disruption of the epithelial-specific Nrf2 pathway increased the severity of eosinophilic sinonasal inflammation.63 In myeloid cell–specific Keap1-deficient mice model, activation of Nrf2 signaling in macrophages and neutrophils improves host survival by increasing antibacterial and anti-inflammatory defenses during sepsis.64 Therefore, Nrf2 activators have pivotal immunomodulating effects on leukocytes to control bacterial infection and protect against sepsis.
The pharmacologic activators of Nrf2 can be classified as electrophiles, protein–protein interaction inhibitors, and multitarget drugs.56 The most common Nrf2 activators are electrophilic molecules that covalently oxidated or alkylated cysteine residues present in the thiol-rich Keap1 protein.65–67 Protein–protein interaction inhibitors activate Nrf2 by interrupting the docking of Nrf2 to the Kelch propeller of Keap1, which exhibits better selectivity than electrophilic compounds.68 Moreover, Nrf2 can be upregulated through Keap1-independent posttranslational modifications (eg, phosphorylation and acetylation). Several kinases have been linked to ARE activation, including extracellular signal-regulated kinase (ERK1/2),69,70 p38 mitogen-activated protein (MAP) kinase,71 glycogen synthase kinase 3 (GSK-3),40 phosphatidylinositol 3-kinase and its downstream target Akt/protein kinase B (PKB), and protein kinase C (PKC).72 Future studies should clarify which mechanisms regulate Nrf2 activation in AuNDs-NH2-treated cells. AuNDs-CH3 did not affect intracellular ROS levels, but it may phosphorylate P62 Ser349 to activate the noncanonical P62–Keap1–Nrf2 pathway. The deficiency in autophagy upregulated P62, thereby inhibiting the Keap1-Cul3-E3 ubiquitin ligase complex and stabilizing Nrf2.73 AuNDs-CH3 affected Nrf2 activation and downstream genes but did not have the obvious phagocytosis ability of AuNDs-NH2. Additional experiments are warranted to determine whether AuNDs-CH3 blocks autophagosome maturation and P62 accumulation, resulting in prolonged Nrf2 activation in a P62-concentration-dependent manner in macrophage. AuNDs-OH also did not affect intracellular ROS levels or activate the Nrf2 pathway, probably due to its surface modifications or insufficient amount entered the cells. Our result indicated that nanozymes not only exhibited intrinsic enzyme-like activity but also triggered intracellular signaling pathways in macrophages through modifications of superficial atoms of nanomaterials. However, future studies should investigate how outfacing groups of primary amine and quaternary ammonium ions in AuNDs phosphorylate different intracellular regulators (such as Nrf2 and P62), leading to different consequences including the expression of cytokine and phagocytic ability in macrophages.
Conclusions
Nrf2 can be a therapeutic target for chronic diseases, such as inflammation and cardiovascular diseases. Preclinical studies have reported that Nrf2 activation has potent anti-inflammatory effects in myeloid leukocytes and macrophages. To the best of our knowledge, this is the first study demonstrating that Nrf2 and its downstream targets are induced in macrophages by AuNDs-NH2 treatment without triggering excess oxidative stress. This finding may provide a rationale strategy for nanomaterials targeting the Nrf2 pathway against certain chronic inflammatory diseases.
Funding
This study was supported by grants MOST-104-2320-B-400-003-MY3 and MOST-108-2320-B-008-001 from the Ministry of Science and Technology in Taiwan, and National Health Research Institutes, Taiwan, ROC (grant number NHRI-108EMPP02).
Disclosure
All authors declare that they have no competing financial interests. The authors report no conflicts of interest for this work.
References
1. Luo YH, Chang LW, Lin P. Metal-based nanoparticles and the immune system: activation, inflammation, and potential applications. Biomed Res Int. 2015;2015:143720. doi:10.1155/2015/143720
2. Brune B, Dehne N, Grossmann N, et al. Redox control of inflammation in macrophages. Antioxid Redox Signal. 2013;19(6):595–637. doi:10.1089/ars.2012.4785
3. Bryan HK, Olayanju A, Goldring CE, Park BK. The Nrf2 cell defence pathway: keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol. 2013;85(6):705–717. doi:10.1016/j.bcp.2012.11.016
4. Thimmulappa RK, Lee H, Rangasamy T, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116(4):984–995. doi:10.1172/JCI25790
5. Kobayashi EH, Suzuki T, Funayama R, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi:10.1038/ncomms11624
6. Dumont M, Wille E, Calingasan NY, et al. Triterpenoid CDDO-methylamide improves memory and decreases amyloid plaques in a transgenic mouse model of Alzheimer’s disease. J Neurochem. 2009;109(2):502–512. doi:10.1111/j.1471-4159.2009.05970.x
7. Kaidery NA, Banerjee R, Yang L, et al. Targeting Nrf2-mediated gene transcription by extremely potent synthetic triterpenoids attenuate dopaminergic neurotoxicity in the MPTP mouse model of Parkinson’s disease. Antioxid Redox Signal. 2013;18(2):139–157. doi:10.1089/ars.2011.4491
8. Yang L, Calingasan NY, Thomas B, et al. Neuroprotective effects of the triterpenoid, CDDO methyl amide, a potent inducer of Nrf2-mediated transcription. PLoS One. 2009;4(6):e5757. doi:10.1371/journal.pone.0005757
9. Boutten A, Goven D, Artaud-Macari E, Boczkowski J, Bonay M. NRF2 targeting: a promising therapeutic strategy in chronic obstructive pulmonary disease. Trends Mol Med. 2011;17(7):363–371. doi:10.1016/j.molmed.2011.02.006
10. Cuadrado A, Rojo AI, Wells G, et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov. 2019;18(4):295–317.
11. Boisselier E, Astruc D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem Soc Rev. 2009;38(6):1759–1782. doi:10.1039/b806051g
12. Bahadar H, Maqbool F, Niaz K, Abdollahi M. Toxicity of nanoparticles and an overview of current experimental models. Iran Biomed J. 2016;20(1):1–11.
13. Luo YH, Wu ZW, Tsai HT, Lin SY, Lin PP. Endotoxin nanovesicles: hydrophilic gold nanodots control supramolecular lipopolysaccharide assembly for modulating immunological responses. Nano Lett. 2015;15(10):6446–6453. doi:10.1021/acs.nanolett.5b01809
14. Liu CP, Wu TH, Lin YL, Liu CY, Wang S, Lin SY. Tailoring enzyme-like activities of gold nanoclusters by polymeric tertiary amines for protecting neurons against oxidative stress. Small. 2016;12(30):4127–4135. doi:10.1002/smll.201503919
15. Jao YC, Chen MK, Lin SY. Enhanced quantum yield of dendrimer-entrapped gold nanodots by a specific ion-pair association and microwave irradiation for bioimaging. Chem Commun (Camb). 2010;46(15):2626–2628. doi:10.1039/b926364k
16. Glushko V, Thaler MS, Karp CD. Pyrene fluorescence fine structure as a polarity probe of hydrophobic regions: behavior in model solvents. Arch Biochem Biophys. 1981;210(1):33–42. doi:10.1016/0003-9861(81)90160-0
17. Pistolis G, Malliaris A, Paleos CM, Tsiourvas D. Study of poly(amidoamine) starburst dendrimers by fluorescence probing. Langmuir. 1997;13(22):5870–5875. doi:10.1021/la970529i
18. Richter-Egger DL, Landry JC, Tesfai A, Tucker SA. Spectroscopic investigations of polyamido amine starburst dendrimers using the solvatochromic probe phenol blue. J Phys Chem A. 2001;105(28):6826–6833. doi:10.1021/jp0100396
19. Chiang HC, Chao HR, Li LA, Tsai FY, Tsou TC. Reliable and sensitive adenovirus-based reporter system for high-throughput screening of dioxins. Environ Technol Inno. 2015;4:8–16. doi:10.1016/j.eti.2015.03.001
20. Chao HR, Wang YF, Chen HT, et al. Differential effect of arecoline on the endogenous dioxin-responsive cytochrome P450 1A1 and on a stably transfected dioxin-responsive element-driven reporter in human hepatoma cells. J Hazard Mater. 2007;149(1):234–237. doi:10.1016/j.jhazmat.2007.07.022
21. Chou WC, Hsu CY, Ho CC, et al. Development of an in vitro-based risk assessment framework for predicting ambient particulate matter-bound polycyclic aromatic hydrocarbon-activated toxicity pathways. Environ Sci Technol. 2017;51(24):14262–14272. doi:10.1021/acs.est.7b02002
22. Lin CH, Yang MH, Chang LW, et al. Cd/Se/Te-based quantum dot 705 modulated redox homeostasis with hepatotoxicity in mice. Nanotoxicology. 2011;5(4):650–663. doi:10.3109/17435390.2010.539712
23. Yeh LC, Chen SP, Liao FH, Wu TH, Huang YT, Lin SY. The bioactive core and corona synergism of quantized gold enables slowed inflammation and increased tissue regeneration in wound hypoxia. Int J Mol Sci. 2020;21:5. doi:10.3390/ijms21051699
24. Liao FH, Wu TH, Huang YT, et al. Subnanometer gold clusters adhere to lipid a for protection against endotoxin-induced sepsis. Nano Lett. 2018;18(5):2864–2869. doi:10.1021/acs.nanolett.7b05464
25. Liao FH, Wu TH, Yao CN, et al. A supramolecular trap to increase the antibacterial activity of colistin. Angew Chem Int Ed Engl. 2020;59(4):1430–1434. doi:10.1002/anie.201912137
26. Liu CP, Wu TH, Liu CY, Cheng HJ, Lin SY. Interactions of nitroxide radicals with dendrimer-entrapped Au8-clusters: a fluorescent nanosensor for intracellular imaging of ascorbic acid. J Mater Chem B. 2015;3(2):191–197. doi:10.1039/C4TB01657B
27. Laibinis PE, Whitesides GM, Allara DL, Tao YT, Parikh AN, Nuzzo RG. Comparison of the structures and wetting properties of self-assembled monolayers of normal-alkanethiols on the coinage metal-surfaces, Cu, Ag, Au. J Am Chem Soc. 1991;113(19):7152–7167. doi:10.1021/ja00019a011
28. Al-Sawaf O, Clarner T, Fragoulis A, et al. Nrf2 in health and disease: current and future clinical implications. Clin Sci. 2015;129(12):989–999.
29. Vara D, Pula G. Reactive oxygen species: physiological roles in the regulation of vascular cells. Curr Mol Med. 2014;14(9):1103–1125. doi:10.2174/1566524014666140603114010
30. Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10(8):1343–1374. doi:10.1089/ars.2007.1957
31. Pham-Huy LA, He H, Pham-Huy C. Free radicals, antioxidants in disease and health. Int J Biomed Sci. 2008;4(2):89–96.
32. Fu PP, Xia Q, Hwang HM, Ray PC, Yu H. Mechanisms of nanotoxicity: generation of reactive oxygen species. J Food Drug Anal. 2014;22(1):64–75. doi:10.1016/j.jfda.2014.01.005
33. Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280(21):20340–20348.
34. Nair S, Doh ST, Chan JY, Kong AN, Cai L. Regulatory potential for concerted modulation of Nrf2- and Nfkb1-mediated gene expression in inflammation and carcinogenesis. Br J Cancer. 2008;99(12):2070–2082.
35. Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116(20):4338–4348.
36. Yang M, Yao Y, Eades G, Zhang Y, Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat. 2011;129(3):983–991. doi:10.1007/s10549-011-1604-1
37. Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem. 2011;286(47):40725–40733. doi:10.1074/jbc.M111.275495
38. Li N, Muthusamy S, Liang R, Sarojini H, Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev. 2011;132(3):75–85. doi:10.1016/j.mad.2010.12.004
39. Li Y, Paonessa JD, Zhang Y. Mechanism of chemical activation of Nrf2. PLoS One. 2012;7(4):e35122. doi:10.1371/journal.pone.0035122
40. Rojo AI, Medina-Campos ON, Rada P, et al. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: role of glycogen synthase kinase-3. Free Radic Biol Med. 2012;52(2):473–487. doi:10.1016/j.freeradbiomed.2011.11.003
41. Jiang T, Harder B, de la Vega MR, Wong PK, Chapman E, Zhang DD. p62 links autophagy and Nrf2 signaling. Free Radical Bio Med. 2015;88:199–204. doi:10.1016/j.freeradbiomed.2015.06.014
42. Singh A, Venkannagari S, Oh KH, et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem Biol. 2016;11(11):3214–3225. doi:10.1021/acschembio.6b00651
43. Prawan A, Kundu JK, Surh YJ. Molecular basis of heme oxygenase-1 induction: implications for chemoprevention and chemoprotection. Antioxid Redox Signal. 2005;7(11–12):1688–1703. doi:10.1089/ars.2005.7.1688
44. Ishii Y, Itoh K, Morishima Y, et al. Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J Immunol. 2005;175(10):6968–6975. doi:10.4049/jimmunol.175.10.6968
45. Yoh K, Itoh K, Enomoto A, et al. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001;60(4):1343–1353. doi:10.1046/j.1523-1755.2001.00939.x
46. Jhang JJ, Yen GC. The role of Nrf2 in NLRP3 inflammasome activation. Cell Mol Immunol. 2017;14(12):1011–1012.
47. Zhao CC, Gillette DD, Li XH, Zhang ZB, Wen HT. Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J Biol Chem. 2014;289(24):17020–17029. doi:10.1074/jbc.M114.563114
48. Wruck CJ, Streetz K, Pavic G, et al. Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J Biol Chem. 2011;286(6):4493–4499. doi:10.1074/jbc.M110.162008
49. Harvey CJ, Thimmulappa RK, Sethi S, et al. Targeting Nrf2 signaling improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model. Sci Transl Med. 2011;3(78):78ra32.
50. Manke A, Wang L, Rojanasakul Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res Int. 2013;2013:942916. doi:10.1155/2013/942916
51. Cuadrado A, Manda G, Hassan A, et al. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol Rev. 2018;70(2):348–383. doi:10.1124/pr.117.014753
52. Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi:10.1146/annurev.pharmtox.46.120604.141046
53. Katoh Y, Iida K, Kang MI, et al. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch Biochem Biophys. 2005;433(2):342–350. doi:10.1016/j.abb.2004.10.012
54. Pauwels EK, Erba PA, Kostkiewicz M. Antioxidants: a tale of two stories. Drug News Perspect. 2007;20(9):579–585. doi:10.1358/dnp.2007.20.9.1162242
55. Cores A, Piquero M, Villacampa M, Leon R, Menendez JC. NRF2 regulation processes as a source of potential drug targets against neurodegenerative diseases. Biomolecules. 2020;10(6):904. doi:10.3390/biom10060904
56. Robledinos-Anton N, Fernandez-Gines R, Manda G, Cuadrado A. Activators and inhibitors of NRF2: a review of their potential for clinical development. Oxid Med Cell Longev. 2019;2019:1–20. doi:10.1155/2019/9372182
57. Williamson TP, Johnson DA, Johnson JA. Activation of the Nrf2-ARE pathway by siRNA knockdown of Keap1 reduces oxidative stress and provides partial protection from MPTP-mediated neurotoxicity. Neurotoxicology. 2012;33(3):272–279. doi:10.1016/j.neuro.2012.01.015
58. Jazwa A, Rojo AI, Innamorato NG, Hesse M, Fernandez-Ruiz J, Cuadrado A. Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid Redox Signal. 2011;14(12):2347–2360. doi:10.1089/ars.2010.3731
59. Bhullar KS, Rupasinghe HPV. Polyphenols: multipotent therapeutic agents in neurodegenerative diseases. Oxid Med Cell Longev. 2013;2013:1–18. doi:10.1155/2013/891748
60. Rangasamy T, Cho CY, Thimmulappa RK, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114(9):1248–1259. doi:10.1172/JCI200421146
61. Fourtounis J, Wang IM, Mathieu MC, et al. Gene expression profiling following NRF2 and KEAP1 siRNA knockdown in human lung fibroblasts identifies CCL11/Eotaxin-1 as a novel NRF2 regulated gene. Resp Res. 2012;13:92. doi:10.1186/1465-9921-13-92
62. Ishii T, Mann GE. Redox status in mammalian cells and stem cells during culture in vitro: critical roles of Nrf2 and cystine transporter activity in the maintenance of redox balance. Redox Biol. 2014;2:786–794. doi:10.1016/j.redox.2014.04.008
63. Ramanathan M, Tharakan A, Sidhaye VK, Lane AP, Biswal S, London NR. Disruption of sinonasal epithelial Nrf2 enhances susceptibility to rhinosinusitis in a mouse model. Laryngoscope. 2020. doi:10.1002/lary.28884
64. Kong XN, Thimmulappa R, Craciun F, et al. Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am J Resp Crit Care. 2011;184(8):928–938. doi:10.1164/rccm.201102-0271OC
65. Hur W, Gray NS. Small molecule modulators of antioxidant response pathway. Curr Opin Chem Biol. 2011;15(1):162–173. doi:10.1016/j.cbpa.2010.12.009
66. Satoh T, McKercher SR, Lipton SA. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radical Bio Med. 2013;65:645–657. doi:10.1016/j.freeradbiomed.2013.07.022
67. Wilson AJ, Kerns JK, Callahan JF, Moody CJ. Keap calm, and carry on covalently. J Med Chem. 2013;56(19):7463–7476. doi:10.1021/jm400224q
68. Richardson BG, Jain AD, Speltz TE, Moore TW. Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway. Bioorg Med Chem Lett. 2015;25(11):2261–2268. doi:10.1016/j.bmcl.2015.04.019
69. Yu R, Chen C, Mo YY, et al. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem. 2000;275(51):39907–39913. doi:10.1074/jbc.M004037200
70. Yu R, Lei W, Mandlekar S, et al. Role of a mitogen-activated protein kinase pathway in the induction of Phase II detoxifying enzymes by chemicals. J Biol Chem. 1999;274(39):27545–27552. doi:10.1074/jbc.274.39.27545
71. Kang KW, Ryu JH, Kim SG. The essential role of phosphatidylinositol 3-kinase and of p38 mitogen-activated protein kinase activation in the antioxidant response element-mediated rGSTA2 induction by decreased glutathione in H4IIE hepatoma cells. Mol Pharmacol. 2000;58(5):1017–1025. doi:10.1124/mol.58.5.1017
72. Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277(45):42769–42774. doi:10.1074/jbc.M206911200
73. Lau A, Wang XJ, Zhao F, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30(13):3275–3285. doi:10.1128/MCB.00248-10
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.