Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 12
Pri-microRNA-124 rs531564 polymorphism minor allele increases the risk of pulmonary artery hypertension by abnormally enhancing proliferation of pulmonary artery smooth muscle cells
Received 28 October 2015
Accepted for publication 13 January 2016
Published 4 May 2017 Volume 2017:12 Pages 1351—1361
DOI https://doi.org/10.2147/COPD.S99318
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Quanzhong Li, Zongjie Qian, Linqing Wang
Department of Cardiology, The Affiliated Hospital of Guilin Medical University, Guilin, Guangxi, People’s Republic of China
Abstract: MicroRNA-124 (miR-124) has been reported to be downregulated in the cells exposed to hypoxia, which was confirmed in our study. We then used online microRNA target prediction tools to identify GRB2, SMAD5, and JAG1 as the candidate target genes of miR-124, and we next validated GRB2 as a direct gene by using luciferase reporter system. We also established the regulatory relationship between miR-124 and GRB2 by showing the negative linear relationship between GRB2 and miR-124 expression. Furthermore, we investigated the miR-124 and GRB2 expression levels of different genotypes including CC (n=30), GC (n=18), and GG (n=4), which supported the hypothesis that the presence of minor allele (C) of rs531564 polymorphism compromised the expression of miR-124. Meanwhile, we also conducted real-time polymerase chain reaction and Western blot analysis to study the expression of GRB2 among different genotypes or pulmonary artery smooth muscle cells (PASMCs) treated with miR-124 mimics, GRB2 small interfering RNA, and miR-124 inhibitors, respectively, and found that introduction of miR-124 or GRB2 small interfering RNA could reduce the expression of GRB2 and inhibit the proliferation of PASMCs, while miR-124 upregulated the expression of GRB2 and promoted the proliferation of PASMCs. A total of 412 COPD patients with PAH (n=182) or without PAH (n=230) were recruited in this study, and more individuals carrying at least one minor allele of rs531564 were found in the COPD patients with PAH than in those without PAH (odds ratio: 0.61, 95% confidence interval: 0.41–0.91; P=0.166). In conclusion, the presence of rs531564 minor allele may increase the risk of PAH in COPD by reducing miR-124 expression, increasing GRB2 expression, and promoting the proliferation of PASMCs.
Keywords: microRNA-124, rs531564, PAH, COPD, PASMC, proliferation, hypoxia
Introduction
Chronic obstructive pulmonary disease (COPD) is a leading cause of morbidity and mortality worldwide with an increasing prevalence during the past decades.1 One established complication of COPD is the development of pulmonary artery hypertension (PAH). Typically, PAH appears when airflow limitation is severe and is associated with chronic hypoxemia, with the main pathophysiological cause being chronic alveolar hypoxia, although new mechanisms have emerged recently.1 Alveolar hypoxia is probably the most important factor leading to an increased peripheral vascular resistance.2 Acute hypoxia induces in humans, as well as in almost all species of mammals, a rise of peripheral vascular resistance and pulmonary artery pressure that is caused by hypoxic pulmonary vasoconstriction.3
Hypoxia-induced pulmonary hypertension (HPH) is characterized by the sustained narrowing of pulmonary artery lumen and vascular remodeling, which contributes to the morbidity and mortality of adult and pediatric patients with hypoxemia caused by a wide spectrum of lung diseases.4 Vascular structure remodeling plays an essential role in the development and persistent deterioration of HPH.4 Even though new therapies have been developed to treat HPH, few of them are effective to halt the progression of HPH.5 One of the major causes of hypoxia-induced vascular remodeling is the abnormally enhanced proliferation of pulmonary artery smooth muscle cells (PASMCs).6 Therefore, a better understanding of the underlying molecular mechanism is of great significance to develop new methods to halt the deterioration of HPH.
MicroRNAs (miRNAs) are a class of noncoding RNAs of 18–25 nucleotides in length that bind to the 3′ untranslated region (3′-UTR) of target mRNA to regulate the gene expression and, consequently, cause degradation or translational repression.7 miRNAs play important roles in the regulation of gene expression, developmental processes, cell differentiation, proliferation and migration, apoptosis, and stress responses.8 Aberrant miRNA expression is directly related to initiation and progression of many pathophysiological processes, including diabetes mellitus, cancer, and cardiac hypertrophy.9 A recent study also implicate a role of miRNAs in the complex pathology and molecular dysregulation that characterizes PAH.10 Several miRNA target genes, including BMPR2, RhoB, SHP2, NFAT, and Mst1, have been predicted to be highly enriched in PAH-associated pathways, suggesting extensive miRNA-regulated control of this disease.10
A class of miRNAs, known as “hypoximiRs”, is characterized by the altered expression in response to hypoxia, with some of them upregulated (such as miR-210) and the others downregulated (such as miR-124).11 The focus of the present study was miR-124, and we confirmed the downregulation of miR-124 in the samples of COPD complicated with PAH, compared with those without PAH. One single-nucleotide polymorphism (SNP; rs531564, C > G) situated within the “seed sequence” of pri-miR-124 has been shown to compromise the processing and production of mature miRNA, and is associated with a reduced expression of the miRNA,12 which may contribute to various human diseases.12 Herein, we studied the effect of rs531564 on the expression of miR-124, GRB2, the proliferation of PASMCs, as well as the development of PAH.
Materials and methods
Study population and lung tissue samples
Exactly 412 COPD patients with PAH (n=182) or without PAH (n=230) were recruited from the Affiliated Hospital of Guilin Medical University. Among them, resected specimens were available in 62 patients who had received surgery for lung tumor resection, including those with PAH (n=30) or without PAH (n=32). From all participants, 5 mL of peripheral blood was obtained. To evaluate the effect of smoking, we conducted pack-years calculation that indicates the cumulative smoking dose (pack-years = [cigarettes per day/20] × years smoked). The other demographic and clinicopathological characteristics are described in Table 1. The research protocol was approved by the research ethic committee of the Affiliated Hospital of Guilin Medical University and written informed consent was obtained from each participant prior to the study.
Genotyping
Genomic DNA was extracted using DNA extraction kit (Huashun, Shanghai, People’s Republic of China). Genotyping was performed using the TaqMan method with ABI 7900 Real-Time polymerase chain reaction (PCR) system (Thermo Fisher Scientific, Waltham, MA, USA). Controls were included in each plate to ensure accuracy of the genotyping.
Cell culture
PASMCs were either purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) or obtained from the patient samples. The cultured PASMCs were genotyped as wild type (WT; rs531564, CC). We also isolated PASMCs from the patients, which was genotyped as rs531564 GC. The PASMCs were maintained in a medium of Roswell Park Memorial Institute 1640 supplemented with 20% fetal bovine serum at 37°C in 5% CO2 (Thermo Fisher Scientific, Waltham, MA, USA) atmosphere. When confluence reached 80%, the cells were trypsinized and split at a ratio of 1:3, and maintained in 5% fetal bovine serum Roswell Park Memorial Institute 1640. Cells of the third to fifth-generation were collected for further experiments. Smooth muscle α-actin was employed to verify smooth muscle cell identity (95% positive of both markers in smooth muscle cells, data not shown).
RNA extraction and reverse-transcription polymerase chain reaction (RT-PCR)
Total mRNA was extracted from the cultured PASMCs or harvested pulmonary artery tissues with Trizol (Thermo Fisher Scientific, Waltham, MA, USA). In order to measure mRNA expression level, an RT-PCR Kit (Qiagen, Valencia, CA, USA) was adopted to perform RT-PCR with 2 mg of total RNA, in accordance with the manufacturer’s protocol. The protocol was conducted as below: 5 minutes at 95°C, then 35 cycles of amplification (denaturation at 95°C for 30 seconds, annealing for 30 seconds [51°C for miR-124, 55°C for β-actin, 59°C for GRB2, 58°C for JAG1, 50°C for U6, and 51°C for SMAD5], extension at 72°C for 40 seconds). Then 1% agarose gel electrophoresis was performed with the polymerase chain reaction products and ethidium bromide staining was carried out to confirm the amplification of the expected molecular size. To calculate the relative expression level of the miRNA and mRNAs, 2−ΔΔCT method was used.
Oligo transfection
PASMCs were seeded in six-well plates (1×106 cells/well) under a normoxia environment. The cells were then cultured under 3% hypoxia for another 24 hours before Western blot or 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. miR-124 mimics/inhibitors and GRB2 small interfering RNA (siRNA) were purchased from Ambion (Austin, TX, USA). Fugene HD transfection reagent was employed to perform the transfection (Hoffman-La Roche Ltd., Basel, Switzerland).
Western blot
The cells were harvested and resuspended with mRIPA mammalian protein extraction lysis buffer (1 mM ethylenediaminetetraacetic acid, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5). The protein extracts were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated protein blots were then transferred to nitrocellulose membrane. The membranes were blocked with 5% fat-free milk in Tris-Tween-Buffer-Saline (0.1% Tween 20, 150 mM NaCl, and 20 mM Tris-HCl, pH 7.6). The corresponding primary antibodies (anti-GRB2 antibody, 1:1,000 [Santa Cruz Biotechnology Inc., Dallas, TX, USA]; anti-β-actin antibody, 1:10,000 [Santa Cruz Biotechnology Inc.]) were used to incubate the blots, followed by horseradish peroxidase-conjugated secondary antibodies at a final concentration of 1:10,000. Enhanced chemiluminescence kit was adopted to develop the membrane before exposing to X-ray film (Thermo Fisher Scientific).
Cell proliferation assay
The MTT method was applied to perform the cell proliferation assay. In short, 0.5 mg/mL MTT was used to incubate PASMCs at 37°C for 4 hours after being treated under hypoxia or normoxia for 48 hours. Subsequently, we removed the culture medium, and used 200 μL dimethylsulfoxide to dissolve the formazan salt crystals and shook them for 10 minutes. Spectramax M2 microplate Reader was employed to read the absorbance at 570 nm (Molecular Devices LLC, Sunnyvale, CA, USA). We performed the trypan blue exclusion in parallel, in order to confirm the validation of the MTT assay.
Hypoxia exposure
PASMCs were exposed to hypoxia provided by connecting to a chamber equilibrated with a water-saturated gas mixture of 1% O2, 5% CO2, and 94% N2.
5-Bromo-2-deoxyuridine incorporation assays
At the end of culture, cells were incubated with goat serum and anti-5-bromo-2-deoxyuridine antibody overnight at 4°C, followed by incubation with biotinylated goat anti-mouse immunoglobulin (Ig)G (1:200) for 1 hour, prior to staining with 3,3′-diaminobenzidine. The percentage of stained cells was calculated by counting the number of positively stained cells divided by the total number of cells in the same vision field.
Plasmid construction and luciferase reporter assay
The 3′-UTRs of GRB2, SMAD5, and JAG1, which have response element of miR-124, were cloned into the pIS0 control luciferase vector (Promega Corporation, Fitchburg, WI, USA). We used site-directed mutagenesis kit (Stratagene) to introduce mutation to each 3′-UTR. PASMC cells were seeded in triplicate in 24-well plates (1×105/well) and cotransfected with WT/mutant (Mut) 3′-UTR vectors and miR-124 mimics/negative control and Renilla luciferase control vector. After 48 hours of transfection, a dual luciferase reporter assay system was applied to detect luciferase activity in accordance with the protocol of the manufacturer (Promega Corporation). When the confluence reached 80%, expressing plasmid of miR-10b and firefly luciferase reporter gene constructs were cotransfected into the cells for 48 hours. A dual luciferase reporter assay system was applied to detect luciferase activity in accordance with the protocol of the manufacturer (Promega Corporation).
Statistical analysis
Categorical variables were analyzed using contingency tables. For continuous variables, median and range were computed. To investigate their relationship with the clinical, pathologic, and biologic parameters, single factor analysis was performed for categorical variables using Pearson’s chi-square test or Fisher’s exact test, when applicable. The relationship between the polymorphism and each patient’s clinicopathologic characteristics was also studied, and regression analyses were carried out to estimate the effect of miR-124 rs531564 polymorphisms on the risk of PAH in the presence of other known prognostic factors, including age, sex, and smoking status. The difference among groups was determined using Student’s t-test (two groups) or one-way analysis of variance (more than two groups). Two-tailed P<0.05 was considered to indicate statistical significance. All statistical analyses were performed on the Statistical Package for the Social Sciences 19.0 software (IBM Corporation, Armonk, NY, USA).
Results
miR-124 was differentially expressed in PASMCs
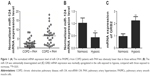
To study the molecular mechanism underlying the development of PAH in patients with COPD, we determined miRNA expressions in COPD sample cells as compared with those without PAH. As shown in Figure 1A, the normalized mRNA expression level of miR-124 in PASMCs from COPD with PAH was obviously lower than that of the controls, indicating the differential expression of miR-124 might be involved in the pathogenesis of PAH in patients with COPD. Furthermore, we cultured PASMCs and exposed the cells to hypoxia, and found that miR-124 was substantially downregulated (Figure 1B) and GRB2 mRNA expression was markedly upregulated (Figure 1C) in the cells exposed to hypoxia, compared with those exposed to normoxia.
GRB2 was a direct target of miR-124
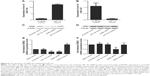
As shown in Figure 2, to identify the candidate target of miR-124, we used online miRNA target prediction tools (www.mirdb.org) and, consequently, identified GRB2, SMAD5, and JAG1 to be the possible target genes of miR-124 with different specific sites on 3′-UTR of each gene. To further identify the exact target gene of miR-124, we treated the PASMCs with constructs containing the WT or Mut target gene 3′-UTR segments together with miR-124 mimics or scramble controls. As shown in Figure 3, cells transfected with WT GRB2 3′-UTR segments showed substantially lower luciferase activity when compared with cells treated with Mut GRB2 3′-UTR segments as well as the scramble controls, while those cells treated with 3′-UTRs of SMAD5 or JAG1 had minimal effect on the luciferase activity. The earlier results all indicated GRB2 is direct target gene of miR-124.
  | Figure 2 GRB2, SMAD5, and JAG1 were identified as candidate target genes of miR-124. |
Furthermore, to explore the regulatory relationship between miR-124 and GRB2, we also investigated the correlation line between GRB2 mRNA and miR-124 expression level. As shown in Figure 4, the expression of GRB2 is correlated with miR-124 in our samples, which revealed the negative regulatory relationship between miR-124 and GRB2.
Expression level of miR-124 and GRB2 was associated with genotype groups of rs531564 polymorphism
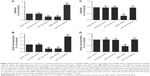
The rs531564 polymorphism was previously reported to be interfering with the expression of miR-124.12 Among the COPD samples collected as shown in Figure 5A, GG (n=30) showed substantially higher miR-124 expression level when compared with GC (n=18) and CC (n=4), indicating that the presence of minor allele (C) of rs531564 polymorphism compromised the expression of miR-124.
We further conducted real-time polymerase chain reaction and Western blot analysis to study the mRNA and protein expression level of GRB2 among different genotypes. As shown in Figure 5B and C, both the mRNA and protein expression level of GRB2 in the GG sample group were substantially lower when compared with the minor allele carrying groups, GC and CC sample groups, indicating the negative regulatory relationship between miR-124 and GRB2.
miR-124 inhibits the expression of GRB2 in PASMCs with CC, but not in PASMCs with GC
To further validate the hypothesis of the negative regulatory relationship between miR-124 and GRB2, we treated PASMCs genotyped as CC with miR-124 mimics, GRB2 siRNA, and miR-124 inhibitors. The effect of miR-124 mimics and inhibitor on its expression was evaluated (Figure 6A and B). As shown in Figure 6C and E, the GRB2 protein (Figure 6A) and mRNA expression level (Figure 6C) of cells (CC) treated with miR-124 mimics or GRB2 siRNA were substantially lower than the scramble control, while cells (CC) treated with miR-124 inhibitors showed substantially higher GRB2 mRNA and protein expression level than the scramble control.
As shown in Figure 6D and F, the GRB2 protein (Figure 6D) and mRNA expression level (Figure 6F) of PASMCs genotyped as GC treated with GRB2 siRNA were substantially lower than the scramble control, while PASMCs genotyped as GC treated with miR-124 mimics or inhibitors did not show any change in the expression of GRB2.
miR-124 interfered with the viability of PASMCs with CC, but not in PASMCs with GC
We also investigated the viability of PASMCs when transfected with miR-124 mimics, GRB2 siRNA, and miR-124 inhibitors, compared with the scramble controls. As shown in Figure 7A and B, cells transfected with miR-124 mimics and GRB2 siRNA showed comparably lower viability when compared with the scramble controls, while cells transfected with miR-124 inhibitors showed substantially higher viability, indicating miR-124 negatively interfered with the viability of PASMCs by targeting GRB2.
As shown in Figure 7C and D, cells transfected with miR-124 mimics and GRB2 siRNA showed comparably lower viability when compared with the scramble controls, while cells transfected with miR-124 inhibitors showed substantially higher viability, indicating miR-124 negatively interfered with the viability of PASMCs by targeting GRB2.
rs531564 is associated with risk of PAH in patients with COPD
A total of 412 COPD patients with PAH (n=182) or without PAH (n=230) were recruited in this study. Information on the known risk factors of PAH, such as age, height, weight, and smoking status, and lung function such as forced vital capacity, forced expiratory volume in 1 second, and diffusion capacity of the lungs for carbon monoxide are described in Table 1. The genotype frequency of the SNP in the study population was compatible with Hardy–Weinberg equilibrium, and the recruited COPD patients showed expected physiologic alterations including significantly lower forced expiratory volume in 1 second and lower diffusion capacity of the lungs for carbon monoxide. Furthermore, by using regression analysis, more individuals carrying at least one minor allele of rs531564 were noted in the COPD patients with PAH that those without PAH (odds ratio: 0.61, 95% confidence interval: 0.41–0.91; P=0.0166).
Discussion
PAH, characterized by increased pulmonary arterial pressure and pulmonary vascular resistance, is a life-threatening disease.13 Even though the exact mechanisms responsible for the development of PAH remains elusive, it has been found that abnormally enhanced proliferation of PASMCs was detected at the early stage of the disease, and numerous factors are involved in the aberrant proliferation of PASMCs.14 Hypoxia has been used as a model of pulmonary hypertension for many years and is a real-life cause of pulmonary hypertension in humans.15 Remodeling of hypoxic pulmonary vasculature results in enhanced muscularization of pulmonary artery. A consensus has been reached that hypoxia could lead to vascular remodeling and proliferation of pulmonary vascular cells. A previous study have demonstrated that hypoxia increases cell proliferation by inducing production and releasing different inflammatory mediators (eg, MCF-1, interleukin-8, interleukin-6) and mitogenic stimuli (eg, vascular endothelial growth factor, serotonin, platelet-derived growth factor, endothelin-1, 5-hydroxytryptamine) from vascular smooth muscle cells and/or by inhibiting antimitogenic factors (eg, prostacyclin and nitric oxide).15 It has been previously reported that miR-124 is downregulated in response to hypoxia exposure,11 and in this study, we confirmed the downregulation of miR-124 in the samples of COPD complicated with PAH, compared with those without PAH. Furthermore, we found that the normalized mRNA expression level of miR-124 in PASMCs from COPD with PAH was obviously lower than that of controls, indicating the differential expression of miR-124 might be involved in the pathogenesis of PAH in patients with COPD.
The miR-124 was initially described as a brain-specific miRNA that is associated with various nervous system diseases such as Alzheimer disease and medulloblastoma.16 However, several reports have shown that miR-124 is also expressed in non-neuron tissues/cells such as pancreas,17 breast,18 bone marrow-derived mesenchymal stem cells,19 lung,20 fibroblast,21 and fetal tissues22 and has functions in these tissues. PASMCs in the adult are highly specialized cells that regulate the vessel tone through the expression of a unique repertoire of SMC-specific genes such as -SMA, SM22, smooth muscle myosin heavy chain (SM-MHC), and calponin.23 Unlike terminally differentiated muscle cells, PASMCs are remarkably plastic and can modulate their phenotype from contractile to proliferative, synthetic type in response to vascular injury.23 Reduced miR-124 expression occurs on acquisition of a highly proliferative, invasive phenotype.24 Downregulation of miR-124 has also been documented in carcinogenesis in other cell systems25 and in rheumatoid arthritis where altered expression of miRNAs, in general, and miR-124 specifically has been shown to regulate cell proliferation and inflammation in synovial fibroblasts.26
Studies in cancer and neuronal cells have begun to establish the downstream targets of miR-124, which may be associated with proliferation or other aspects of activated cell phenotype, such as Sox9, small C-terminal domain phosphatase 1, polypyrimidine tract–binding protein 1 (PTBP1), Jag1, ephrin-B1, NfatC3, and monocyte chemotactic protein-1.27–29 PTBP1 is especially interesting because of its effects on notch signaling and other aspects of proliferation in cancer cell types.30 There are three cell types including smooth muscle cells, fibroblasts, and endothelial cells that are involved in the pathogenesis of PAH. PTBP1 contributes to the disease by its effect on fibroblasts,30 and one miRNA may have different target genes in different cell types. In this study, we identified GRB2, SMAD5, and JAG1 as the possible targets of miR-124 and validated GRB2 as a direct target of miR-124 by using luciferase assay. In addition, we showed that the expression of GRB2 is correlated with miR-124 in human tissue samples, and the GRB2 protein and mRNA expression level of cells treated with miR-124 mimics or GRB2 siRNA were substantially lower than those of scramble control, while cells treated with miR-124 inhibitors showed substantially higher GRB2 mRNA and protein expression level than those of scramble control.
The expression of GRB2 has been detected in a wide range of tissues, and it plays a key role in regulating multiple cellular functions.31 Inhibition of GRB2 function impairs developmental processes in various organisms and suppresses the transformation and proliferation of various cell types.32 GRB2 is well known for its role to link the epidermal growth factor receptor tyrosine kinase to the activation of Ras and its downstream kinases, which may explain its ability to enhance the proliferation of human cells.33 Evidence demonstrates that ERK2/GRB2/Shc pathway is critical in vascular smooth muscle cell proliferation which is mediated by c-Src, though the precise signaling mechanisms by which c-Src is involved in critical cellular events remain opaque.34 It has been reported that MEK kinase phosphorylates ERK1/2 upon mitogenic stimulation, followed by entry into the nucleus, where DNA synthesis and expression of cell cycle regulators are regulated.35 In this study, we found that the cells transfected with miR-124 mimics and GRB2 siRNA showed comparably lower viability when compared with the scramble controls, while cells transfected with miR-124 inhibitors showed substantially higher viability, indicating miR-124 negatively interfered with the viability of PASMCs by targeting GRB2.
One SNP (rs531564, C > G) situated within the “seed sequence” of pri-miR-124 has been shown to compromise the processing and production of mature miRNA, and is associated with a reduction in expression of the miRNA.12 In this study, we collected tissue samples and genotyped them as GG (n=30), GC (n=18), and CC (n=4), and GG genotype group showed substantially higher miR-124 expression level when compared with GC or CC, whereas both the mRNA and protein expression level of GRB2 in the GG sample group were substantially lower when compared with the minor allele carrying groups, GC and CC sample groups. Furthermore, we collected blood samples from 412 COPD patients with (n=182) or without (n=230) PAH and found that more individuals carrying at least one minor allele of rs531564 were noted in the COPD patients with PAH than in those without PAH (odds ratio: 0.61, 95% confidence interval: 0.41–0.91; P=0.166) by using regression analysis.
Conclusion
In conclusion, our findings presented a novel role of miR-124 in repressing the proliferation of PASMCs by targeting GRB2, providing an important lead for the development of PAH in response to hypoxia. The presence of rs531564 minor allele may increase the risk of PAH in COPD by reducing miR-124 expression, increasing GRB2 expression, and promoting the proliferation of PASMCs.
Acknowledgment
This research was supported by China Natural Science Foundation (81460052) and Guangxi Natural Science Foundation (2014GXNSFAA118153).
Disclosure
The authors report no conflicts of interest in this work.
References
Celli BR, MacNee W, ATS/ERS Task Force. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23(6):932–946. | ||
Fishman AP. Hypoxia on the pulmonary circulation. How and where it acts. Circ Res. 1976;38(4):221–231. | ||
Fishman AP, McClement J, Himmelstein A, Cournand A. Effects of acute anoxia on the circulation and respiration in patients with chronic pulmonary disease studied during the steady state. J Clin Invest. 1952;31(8):770–781. | ||
Stenmark KR, Fagan KA, Frid MG. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ Res. 2006;99(7):675–691. | ||
Zakynthinos E, Daniil Z, Papanikolaou J, Makris D. Pulmonary hypertension in COPD: pathophysiology and therapeutic targets. Curr Drug Targets. 2011;12(4):501–513. | ||
Pak O, Aldashev A, Welsh D, Peacock A. The effects of hypoxia on the cells of the pulmonary vasculature. Eur Respir J. 2007;30(2):364–372. | ||
Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Bauersachs J, Thum T. Biogenesis and regulation of cardiovascular microRNAs. Circ Res. 2011;109(3):334–347. | ||
Parikh VN, Jin RC, Rabello S. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: results of a network bioinformatics approach. Circulation. 2012;125(12):1520–1532. | ||
Greco S, Martelli F. MicroRNAs in hypoxia response. Antioxid Redox Signal. 2014;21(8):1164–1166. | ||
Gao XR, Wang HP, Zhang SL, Wang MX, Zhu ZS. Pri-miR-124 rs531564 polymorphism and colorectal cancer risk. Sci Rep. 2015;5: 14818. | ||
Badesch DB, Champion HC, Sanchez MA, et al. Diagnosis and assessment of pulmonary arterial hypertension. J Am Coll Cardiol. 2009; 54(1 Suppl):S55–S66. | ||
Rubin LJ. Pulmonary arterial hypertension. Proc Am Thorac Soc. 2006; 3(1):111–115. | ||
Welsh DJ, Peacock AJ. Cellular responses to hypoxia in the pulmonary circulation. High Alt Med Biol. 2013;14(2):111–116. | ||
Agirre X, Vilas-Zornoza A, Jiménez-Velasco A, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69(10):4443–4453. | ||
Baroukh N, Ravier MA, Loder MK, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282(27):19575–19588. | ||
Bierer R, Nitta CH, Friedman J, et al. NFATc3 is required for chronic hypoxia-induced pulmonary hypertension in adult and neonatal mice. Am J Physiol Lung Cell Mol Physiol. 2011;301(6):L872–L880. | ||
Laine SK, Alm JJ, Virtanen SP, Aro HT, Laitala-Leinonen TK. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2012;113(8):2687–2695. | ||
Catuogno S, Cerchia L, Romano G, Pognonec P, Condorelli G, de Franciscis V. miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene. 2013;32(3):341–351. | ||
Wang D, Zhang H, Li M, et al. MicroRNA124 controls the proliferative, migratory, and inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res. 2014;114(1):67–78. | ||
Wang Y, Huang C, Chintagari NR, Xi D, Weng T, Liu L. miR124 regulates fetal pulmonary epithelial cell maturation. Am J Physiol Lung Cell Mol Physiol. 2015;309(4):L400–L413. | ||
Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84(3):767–801. | ||
Zhao WH, Wu SQ, Zhang YD. Downregulation of miR-124 promotes the growth and invasiveness of glioblastoma cells involving upregulation of PPP1R13L. Int J Mol Med. 2013;32(1):101–107. | ||
Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 2013;25(2):215–221. | ||
Nakamachi Y, Kawano S, Takenokuchi M, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–1304. | ||
Kang K, Peng X, Zhang X, et al. MicroRNA-124 suppresses the transactivation of nuclear factor of activated T cells by targeting multiple genes and inhibits the proliferation of pulmonary artery smooth muscle cells. J Biol Chem. 2013;288(35):25414–25427. | ||
Akerblom M, Sachdeva R, Jakobsson J. Functional studies of microRNAs in neural stem cells: problems and perspectives. Front Neurosci. 2012;6:14. | ||
Arvanitis DN, Jungas T, Behar A, Davy A. Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Mol Cell Biol. 2010;30(10):2508–2517. | ||
Cheung HC, Corley LJ, Fuller GN, McCutcheon IE, Cote GJ. Polypyrimidine tract binding protein and Notch1 are independently re-expressed in glioma. Mod Pathol. 2006;19(8):1034–1041. | ||
Levy-Apter E, Finkelshtein E, Vemulapalli V, Li SS, Bedford MT, Elson A. Adaptor protein GRB2 promotes Src tyrosine kinase activation and podosomal organization by protein-tyrosine phosphatase in osteoclasts. J Biol Chem. 2014;289(52):36048–36058. | ||
Li QY, Zhu YF, Zhang M, et al. Chlorogenic acid inhibits hypoxia-induced pulmonary artery smooth muscle cells proliferation via c-Src and Shc/Grb2/ERK2 signaling pathway. Eur J Pharmacol. 2015;751:81–88. | ||
Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. Role of Grb2 in EGF-stimulated EGFR internalization. J Cell Sci. 2002;115(Pt 9):1791–1802. | ||
Sayeski PP, Ali MS. The critical role of c-Src and the Shc/Grb2/ERK2 signaling pathway in angiotensin II-dependent VSMC proliferation. Exp Cell Res. 2003;287(2):339–349. | ||
Adam RM, Borer JG, Williams J, Eastham JA, Loughlin KR, Freeman MR. Amphiregulin is coordinately expressed with heparin-binding epidermal growth factor-like growth factor in the interstitial smooth muscle of the human prostate. Endocrinology. 1999;140(12):5866–5875. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.