Back to Journals » Infection and Drug Resistance » Volume 11
Prevalence of Plasmodium falciparum antimalarial drug resistance genes in Southeastern Gabon from 2011 to 2014
Authors Voumbo-Matoumona DF, Kouna LC , Madamet M, Maghendji-Nzondo S, Pradines B, Lekana-Douki JB
Received 18 December 2017
Accepted for publication 23 February 2018
Published 28 August 2018 Volume 2018:11 Pages 1329—1338
DOI https://doi.org/10.2147/IDR.S160164
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Dominique Fatima Voumbo-Matoumona,1,2,3 Lady Charlène Kouna,4 Marylin Madamet,2,5,6 Sydney Maghendji-Nzondo,4 Bruno Pradines,2,5,6 Jean Bernard Lekana-Douki1,4
1Unit of Evolution, Epidemiology and Parasitic Resistances (UNEEREP), International Medical Research Center of Franceville (CIRMF), Franceville, Gabon; 2Parasitology and Entomology Unit, Department of Infectious Diseases, Biomedical Research Institute of Army, Marseille, France; 3Regional Doctoral School of Central Africa in Tropical Infectiology, Franceville, Gabon; 4Department of Parasitology, Mycology and Tropical Medicine, University of Health Sciences, Libreville, Gabon; 5Research Unit on Infectious and Tropical Emerging Diseases, Aix Marseille University, Marseille, France, 6National Malaria Reference Center, Marseille, France
Purpose: The introduction of artemisinin-based combination therapies (ACTs) in treating uncomplicated malaria and sulfadoxine–pyrimethamine (SP) as intermittent preventive treatment during pregnancy drastically decreased the burden of malarial disease around the world. However, ACTs are known to select for drug resistance markers. In Gabon, artemether–lumefantrine induced an increase in the prevalence of N86-Pfmdr1, which is associated with treatment failure. However, little data are available regarding resistance markers in Southeastern Gabon. This study aimed to evaluate the evolution of resistance haplotypes in the Pfcrt, Pfdhps, Pfdhfr, and PfK13 genes from 2011 to 2014 in Southeastern Gabon.
Methods: A total of 233 Plasmodium falciparum DNA samples were collected from febrile pediatric patients in South Gabon: Franceville, an urban area; Koulamoutou, a semi-urban area; and Lastourville, a rural area. Pfcrt, Pfdhps, Pfdhfr, and the propeller domain of PfK13 were sequenced for all isolates.
Results: The overall prevalence (3.7%–11.5%) of the wild-type haplotype Pfcrt 72-76 CVMNK was not significantly different between 2011 and 2014 in Southeast Gabon. For Pfdhfr (codons 51, 59, 108, 164), the IRNI triple-mutant haplotype was the most prevalent (>89.0%). The ICNI and NCNI mutant haplotypes and the NCSI wild-type haplotype showed a minor prevalence. There were no differences in the distributions of these haplotypes across the 4 years and the three study sites. For Pfdhps, the AAKAA and SGKAA mutant haplotypes and the SAKAA wild-type haplotype were similarly present in the three areas during the study period. The AGKAA double mutant was first observed in 2013 in Franceville and in 2014 in Koulamoutou and Lastourville. Interestingly, only the A578S mutation (0.4%) and two new A494V (0.4%) and V504A (0.9%) mutations were found in PfK13.
Conclusion: Despite the withdrawal of chloroquine, the frequency of the resistant allele 76T remained high in the south of Gabon. Moreover, a high level of resistant haplotypes against IPTp-SP was found.
Keywords: Plasmodium falciparum, antimalarial drug resistance, Pfdhfr/Pfdhps, Pfcrt, PfK13, Gabon
Introduction
Malaria is the most important parasitic disease in the world and affects children and pregnant women in particular. Africa has the most substantial burden, especially sub-Saharan African countries, with ~216 million cases of illness and 445,000 deaths in 2016.1 The emergence of drug resistance remains a major obstacle in the fight against malaria. Given the spread of resistance to the major antimalarial drugs, the World Health Organization (WHO) has recommended the use of artemisinin-based combination therapies (ACTs) to treat uncomplicated malaria and sulfadoxine–pyrimethamine (SP) as an intermittent preventive treatment during pregnancy (IPTp). Consequently, since 2010, the mortality rates due to malaria have fallen by 32%.1 However, the emergence of artemisinin resistance, manifested by delayed parasite clearance after monotherapy with artesunate or ACTs, was described in Southeast Asia.2,3 The withdrawal of previous antimalarial drugs and the switch to ACTs were followed in some countries by a reemergence of some wild-type alleles that were associated with susceptibility to the previous drug treatment. This was reported for the Pfcrt chloroquine marker (Plasmodium falciparum chloroquine resistance transporter) in Tanzania. The K76 wild-type haplotype increased after abandoning chloroquine use and implementing artemether–lumefantrine treatment.4
Two genes, Pfdhfr (P. falciparum dihydrofolate reductase) and Pfdhps (P. falciparum dihydropteroate synthase), have been associated with pyrimethamine and sulfadoxine resistance, respectively, around the world.5 The triple Pfdhfr mutation N51I, C59R, and S108N in combination with the double A437G and K540E Pfdhps mutant formed a quintuple mutant haplotype, which confers a high risk of treatment failure with SP.6 Recent studies have shown an increase in Pfdhps mutations at A581G, thus further escalating the risk for even higher levels of resistance and significant decreases in the effectiveness of SP as IPTp.7,8
The P. falciparum kelch 13 (PfK13) propeller gene has been associated with artemisinin resistance in Southeast Asia since 2013.9 Currently, 17 mutations have been highlighted and associated with artemisinin in Southeast Asia. Among these mutations, the C580Y, Y493H, R539T, and I543T mutations are strongly associated with slow-clearing parasites.2,9–11 Many studies have been conducted in Africa but none of the specific Asian PfK13 mutations associated with artemisinin resistance were found.10,12–17 The A578S mutation was often reported in Africa 10,12,17–20 but all the patients carrying these mutant parasites were cured by artemether–lumefantrine or artesunate-amodiaquine (ASAQ) before day 3.10,12,17–23
In Gabon, a hyperendemic country, the use of ACT as the first line of treatment for uncomplicated malaria has been effective since 2005; whereas ASAQ and artemether–lumefantrine have been the first-line treatment and dihydroartemisinin-piperaquine (DHA-PPQ) has been a second-line treatment for uncomplicated malaria. Severe malaria is treated with injectable quinine or artesunate. Consequently, a significant decrease in the malaria burden has been observed in Libreville and Franceville.24,25 Moreover, SP is used as an IPTp for pregnant women.24 This measure contributed to a significant drop in malaria in pregnant women.26 Chloroquine was the drug used before the introduction of ACT, and its resistance was well described in Gabon. A high level of K76T mutation in Pfcrt was reported.25,27,28 Before the implementation of IPTp with SP, studies reported treatment failures with SP.24,29 During the period of implementation of IPTp with SP in Libreville and Lambarene, a preliminary study reported a high prevalence of multiple mutations in Pfdhfr (nearly 98.0%).24 In North Gabon, 91.9% of Pfdhfr triple mutants and 64.8% of quadruple mutants (Pfdhps A437G) were observed 3 years after this implementation.30 The frequencies of Pfdhfr triple and quintuple mutants (Pfdhps S436A and A437G) increased from 92.9% to 100% and from 17.9% to 75.6%, respectively, between 2005 and 2011 in pregnant women treated after IPTp with SP.31 A recent exploration of PfK13 at Libreville did not detect any mutation.18 There are no available data on the prevalence of PfK13 mutations in South Gabon.
The aim of this retrospective study was to determine the evolution of the prevalence of wild-type/mutant haplotypes for different genes linked to resistance, such as Pfcrt (codons 72 to 76), Pfdhfr (51, 59, 108, 164), Pfdhps (436, 437, 540, 581, 613), and PfK13 between 2011 and 2014 in Lastourville, Koulamoutou, and Franceville, three cities located in Southeast Gabon.
Methods
Study sites
The present study was approved by the Gabonese National Ethics Committee (no 0023/2013/SG/CNE). Parents or guardians gave their written informed consent before collection of blood samples. This study was conducted in three cities located in Southeast Gabon: Franceville, an urban area, from 2011 to 2014; Koulamoutou, a semi-urban area, from 2013 to 2014; and Lastourville, a rural area, from 2013 to 2014 (Figure 1). The transverse collection of peripheral and venous blood in children under 15 years of age was performed.
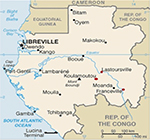  | Figure 1 Localization of study sites: Franceville, Koulamoutou, and Lastourville. |
Diagnosis and DNA extraction
Plasmodial infection was diagnosed using thin blood smears according to Lambarene’s method.31 DNA extraction was done from archived whole blood using the DNA Blood Omega Bio-tek E.Z.N.A® method (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s protocol as previously described.25
Antimalarial resistance gene single-nucleotide polymorphisms (SNPs)
The Pfcrt, Pfdhfr, and Pfdhps genes were amplified by polymerase chain reaction (PCR) using the primers listed in Table 1. The reaction mixture and programs for the thermal cycler were done as previously described.32–35 Briefly, the reaction mixture consisted of Red Diamond Taq DNA buffer (50 mM KCl, 10 mM Tris, pH 8.3), 200 µM deoxynucleotide triphosphate (dNTP), ~200 ng of genomic DNA, 0.32 µM forward and reverse primers, and 0.3 U of Red Diamond Taq DNA polymerase in a final volume of 50 µL. The concentration of MgCl2 was 2.5 mM for Pfdhfr, Pfdhps, and PfK13 (primary PCR), 4.5 mM for Pfcrt and 5 mM for PfK13 (nested PCR). The thermal cycler was programmed differently depending on the gene to be amplified as described previously.33
  | Table 1 Primer sequences and hybridization temperatures Abbreviation: PCR, polymerase chain reaction. |
The PfK13 propeller gene was amplified using a PCR and nested PCR method as described previously.15 Primer sequences and hybridization temperatures are shown in Table 1.
The amplicons were sequenced according to the Sanger sequencing method using 4 μL of BigDye Terminator® v3.1 mix (Life Technologies, Carlsbad, CA, USA) and 0.8 μM of the primers described in Table 1 in a 20 μL volume. The cycle conditions were an initial denaturation at 96°C for 5 min, followed by 30 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Excess dye terminators were removed with a BigDyeXTerminator® Purification Kit (Life Technologies). The samples were loaded on an ABI Prism 3100 analyzer (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturers’ instructions. The sequences were analyzed using the Vector NTI advanced (TM) software (version 11; Thermo Fisher Scientific) to identify specific SNP combinations.
Statistical methods
All data were recorded in Excel (Office 2013). Statistical analysis was carried out with the R software version 3.2.1. Age was expressed as the median and interquartile range (IQR), and parasite densities were expressed as the geometric means (GMPD). The chi-square test was used to compare the categorical variables among the groups. The nonparametric Kruskal–Wallis test and Fisher’s exact test were used for group comparisons, as appropriate. Significance was assumed at p<0.05.
Results
Parasitemia of P. falciparum infected samples
A total of 233 samples infected by P. falciparum were collected from 2011 to 2014 in Franceville, Koulamoutou, and Lastourville (Table 2). There was a significant difference in the comparison of parasitemia among the three localities in 2013. The parasitemia was higher in Franceville and Lastourville (17,022 and 17,754 parasites/µL, respectively) than it was in Koulamoutou (2,905 parasites/µL) (p=0.006). In 2014, the parasitemia was highest in Lastourville (16,586 parasites/µL), followed by Franceville (8,878 parasites/µL) and Koulamoutou (2,192 parasites/µL) (p=0.004). A significant difference was found in the mean ages of the children from the three cities in 2013 and 2014 (Table 2). In 2013, the mean age was highest in Franceville (79 months), followed by Koulamoutou (42 months) and Lastourville (36 months) (p=0.0005). In 2014, the children from Franceville (66 months) and Koulamoutou (50 months) were older than those from Lastourville (36 months) (p=0.007). However, no difference was observed between Franceville and Koulamoutou (p=0.59). Furthermore, no difference was found in each site during the study period (p>0.05).
P. falciparum antimalarial resistance genes
Pfcrt
At least 89.5% of the isolates presented only one Pfcrt polymorphism. CVIET was the main haplotype, and its overall prevalence was stable during the study period: 88.2% (n=15), 96.3% (n=24), 84.7% (n=61), and 88.5% (n=54) from 2011 to 2014, respectively, at the three sites. The overall prevalence of CVMNK, which is associated with chloroquine susceptibility, was also not significantly different over the years: 5.9% (n=1 in 2011), 3.7% (n=1 in 2012), 9.7% (n=7 in 2013), and 11.5% (n=7 in 2014). Mixed genotypes were found only in 2011 (5.9%, n=1) at Franceville and 2013 in all sites (5.4%, n=4).
At Franceville, an increase in the frequency of the CVMNK haplotype was found between 2012 (n=1, 4.0%) and 2014 (n=5, 29.4%) (p=0.03) (Table 3). However, the number of parasites with CVMNK haplotype was too low to conclude that there was a reemergence of chloroquine-susceptible strains. In 2014, the CVMNK frequency in Franceville (n=5, 29.4%) was significantly higher than that in Lastourville (n=1, 5.9%) and Koulamoutou (n=1, 3.7%) (p<0.04). However, there was no significant difference in the frequencies of this haplotype in the other years (p>0.05).
  | Table 3 Prevalence of Pfcrt haplotypes |
Pfdhfr
The analysis of Pfdhfr sequences showed that there were no mixed genotypes in all the isolates, except in one from Franceville in 2014. Only two isolates from Lastourville, one collected in 2013 and one in 2014, harbored the wild-type NCSI haplotype for codons 51, 59, 108, and 164. The prevalence of the IRNI (N51I, C59R, S108N) triple mutants that were associated with pyrimethamine resistance remained high in the three sites. Its overall prevalence was 100% (n=15), 95.7% (n=22), 98.6% (n=73), and 95.2% (n=59) for 2011, 2012, 2013, and 2014, respectively. No I164E mutation was observed in this study.
The NCNI (n=1, 4.3%) and ICNI (n=1, 5.3%) haplotypes were present only in isolates from Franceville in 2012 and 2014, respectively. There was no significant difference in the frequency of each haplotype between the study sites and the years (p≥0.33) (Table 4).
  | Table 4 Prevalence of Pfdhfr and Pfdhps haplotypes Note: Quadruple mutations = Pfdhfr (51+59+108) and Pfdhps (437). |
Pfdhps
Four Pfdhps haplotypes were found from all sites during the study period. There was no mixed genotype. The haplotype harboring only the 436 mutation (AAKAA) was observed with a low frequency of 6.9% and 9.7% in 2013 and 2014, respectively, at all three sites. Globally, the 436/437 (AGKAA) double mutant was present at 1.7% in 2013 (only at Franceville) and 16.1% in 2014 (at all three sites). The haplotype with only the 437 mutation (SGKAA) was the most prevalent in all study sites and all years, at 100% (n=4), 72.4% (n=42), and 64.5% (n=20) for 2011, 2013, and 2014, respectively. No 540E and 581G mutations were observed, and only one isolate from Koulamoutou in 2014 carried the A613S mutation.
There was no significant difference in the frequency of each haplotype between the study sites and the years (p≥0.05) (Table 4).
Pfdhfr–Pfdhps
The overall prevalence of the quadruple mutant (Pfdhps [437] and Pfdhfr [51+59+108]) was 100% (n=3), 80.4% (n=51), and 92.9% (n=28) for 2011, 2013 and 2014, respectively, with no significant increase between 2013 and 2014 (p=1). In each site, the distribution of the quadruple mutant (Pfdhps [436/437] and Pfdhfr [51+59+108]) was not significantly different during the study period at Franceville (p=0.32), Koulamoutou (p=0.32), and Lastourville (p=0.32) (Table 4).
No quintuple (Pfdhps [437+540] and Pfdhfr [51+59+108]) or sextuple (Pfdhps [437+540+581] and Pfdhfr [51+59+108]) mutants were observed.
PfK13
The propeller domain of the PfK13 gene did not show a great diversity of alleles. Of the 233 sequenced isolates, only four carried one of the non-synonymous mutations as follows: A578S (n=1, 0.4%), A504V (n=1, 0.4%), V494A (n=2, 0.9%). Three isolates collected in 2011 in Franceville harbored the A578S, A504V, or V494A mutation. One isolate collected in 2013 in Koulamoutou carried the V494A mutation.
Discussion
To investigate the evolution of antimalarial drug resistance, children younger than 6 and 168 months, with a mean age between 36 and 79 months, were identified at the three sites. We retrospectively analyzed samples from 2011 to 2014, but the data were only from 2013 to 2014 for Lastourville and Koulamoutou.
There was a significant difference in the mean age between 2013 and 2014 at the three sites. The children enrolled in Lastourville and Koulamoutou (rural and semi-urban areas, respectively) were younger than those from the urban site of Franceville. This was consistent with previous studies, which reported the association of an increase of the mean age of P. falciparum-infected children with the drop of malaria prevalence in urban localities, such as Franceville, and in other endemic areas, such as Gambia.36–39 This increase is due to the delay of immunity acquisition. Indeed, the decrease in the malaria burden has led to a decrease in the number of infective bites, which consequently limits immune stimulation. It was previously shown that the malaria prevalence was higher in Lastourville than in Franceville and Koulamoutou, leading to a higher antimalarial immunity in Lastourville than in Franceville and Koulamoutou.37 Indeed, parasitemia was higher in the rural site in 2013 and 2014. A probable hypothesis is the better availability of antimalarial treatment and preventive measures in urban areas. These findings can also be associated with the prevalence of other parasitic or infectious diseases in rural areas that increased the anti-infectious disease immunity, as previously reported.40–43
P. falciparum drug resistance remains a challenge for controlling the malarial burden. The analysis of Pfcrt (chloroquine resistance marker) revealed a significant increase in the chloroquine-susceptible wild-type haplotype CVMNK between 2012 and 2014 in Franceville (the urban area), despite a high level of the resistant haplotype CVIET. However, although the results were significant, these data were determined in a small number of samples (1/17 in 2011, 1/25 in 2012, 2/19 in 2013, and 5/17 in 2014), suggesting an absence of a noticeable reemergence of strains susceptible to chloroquine. The prevalence of resistant strains to chloroquine was still 70.6%. In the semi-urban and rural sites, no significant difference was observed, and the prevalence of the resistant haplotype CVIET remained high since 2000.27 Despite the implementation of ACT and the withdrawal of chloroquine, the frequency of the resistant allele 76T remained high in Franceville (93.8% in 2004, 96% in 2009, and from 70.6% to 96.3% between 2011 and 2014) and in all of Gabon (100% from 1995 until 2002, 97% from 2005 until 2007, and above 70% since 2008).25,27,28,38 Recent studies conducted in Gabon from 1995 to 2008 showed an increased but non-significant amount of the wild-type allele K76, as shown in other studies in Sub-Saharan Africa.27,28,44–46 In South Gabon, the frequency of the resistant allele 76T was higher at 2009.47 There can be a long delay in the selection of the T76 Pfcrt genotype, as shown in previous data from Zanzibar, where this selection was not observed in 2005 but clearly appeared in 2009.4,48 Indeed, recent data from Franceville showed a significant decrease of T76 prevalence around 10 years after chloroquine withdrawal.37 These findings suggest that the selection of T76 Pfcrt takes more time after changes in the antimalarial policy. So, some explanations of the non-significant variation of T76 prevalence between 2011 and 2014 could be the short time and also the smallness of the study effectives here. Furthermore, the T76 prevalence found at 2014 was similar to the previous findings.37 The high frequency of the resistant allele 76T in the south of Gabon may also be due to cross resistance with amodiaquine used in combination with artesunate despite the withdrawal of chloroquine.49–51. On other hand, some reports highlighted that an increase in chloroquine resistance has been observed in recent years following a decrease due to the withdrawal of chloroquine and the introduction of ACT in 2002 in Senegal.
Because of the implementation of IPTp with SP, the Pfdhfr and Pfdhps haplotypes associated with SP resistance were investigated. According to published data from Gabon, the most frequent Pfdhfr haplotype found was the triple-mutant IRNI, which is present in the three localities. A previous study showed that IRNI was the most prevalent Pfdhfr haplotype found both in Lambarene (>92%) in 2007 in Northwest Gabon and in Libreville in 2011 in pregnant women after IPTp with SP (100%).24,29,30,52 These high prevalences of IRNI (>90%) were also observed in Central Africa.53–55 The withdrawal of IPT with SP induced the decline of the IRNI haplotype in North-western Ethiopia, suggesting that the high prevalence of IRNI is maintained due to the use of pyrimethamine.56
Among the successfully analyzed Pfdhps sequence, only mutations at codons 436 and/or 437 have been observed in the three localities. The SGKAA haplotype bearing the 437 single mutation is predominant (50%–100%). These data were consistent with the selection of the 437G mutation reported recently between 2005 and 2008 in North Gabon.30 A non-significant increase in the haplotype with the double mutations 436 and 437 AGKAA has been observed since 2013 in Franceville and since 2014 in Koulamoutou and Lastourville. No haplotype with the 540 and 581 mutations has been observed, contrary to other African countries, such as the Congo, which is a neighbor of Gabon.57 Indeed, the K540E mutation was found in Libreville between 2005 and 2006 and in Lambarene.24,29 However, previous data from the Haut Ogooué province did not report mutations in codons 540 and 581 in 2000.58 The K540E and A581G mutations contributed strongly to reducing the effectiveness of SP during pregnancy.5,7,59 The A613S mutation, which is rarely found in Africa, was present in only one isolate. No A613T mutations described in Asia and East Africa were found in our study.24,60,61
The high prevalence of the IRNI haplotype of Pfdhfr and the SGKAA of Pfdhps concomitantly in urban and rural areas suggested a similar level of adhesion of IPT with SP in urban and rural areas. The absence of high-grade resistance mutant haplotypes (Pfdhfr [51+59+108] + Pfdhps [437+540+581]) associated with IPTp-SP failures during pregnancy in the three localities suggests the efficiency of SP as IPTp.62 This efficiency was associated with a decrease in the prevalence of microscopic P. falciparum infection during pregnancy following IPTp-SP implementation in the urban cities of Gabon. 63 However, the highest level of the triple Pfdhfr mutant plus Pfdhps 437 mutant, considered as marker for SP failure, call for the need of IPTp-SP efficacy monitoring.
Monitoring the emergence and spread of artemisinin resistance is a great challenge. However, the emergence of artemisinin resistance, as manifested by delayed parasite clearance after monotherapy with artesunate or ACT, was described in Southeast Asia.2,3 Several mutations in the PfK13 propeller gene and, more specifically, the C580Y, Y493H, R539T, and I543T mutations were associated with artemisinin resistance in Southeast Asia.2,9–11 None of the mutations described in Southeast Asia that were linked to artemisinin resistance were detected in our samples. The use of bitherapies in the treatment of malaria can delay the apparition of drug resistance. Also, the very short half-life of artemisinin is not favorable for the selection of resistant parasite. These two factors could explain the fact that, less than 10 years after ACT implementation, no molecular markers for artemisinin resistance was observed. Those results were consistent with previous studies in Africa.10,12–17,64 The A578S mutation was detected in one isolate (0.4%). This mutation was often reported in Africa but it had a low prevalence (<2%).10,12,17–20,22,23 This mutation was also identified in Bangladesh.65 However, A578S mutation was not associated with artemisinin treatment failure and all the patients carrying the mutant parasites were cured by artemether–lumefantrine or ASAQ before day 3.10,21 Furthermore, the absence of a role for the A578S mutation in artemisinin resistance was confirmed in an in vitro study using site-directed mutagenesis.10 Two others mutations, V494A (0.4%) and A504V (0.9%), were identified for the first time in two isolates. A non-synonymous mutation V494I was observed in Angola and in Mozambique but was not associated with artemisinin resistance.66 This study described new PfK13 polymorphisms, but the lack of an in vitro assay did not allow us to determine the absence of resistance to artemisinin in the southeast of Gabon. However, the role of PfK13 polymorphisms in artemisinin derivatives in Africa is still debated. Recent publications reported that parasites collected from African patients who were still parasitemic at day 3 and beyond did not carry mutations in PfK13 after ACT treatment.67–70
Conclusion
This study showed a high prevalence of a Pfcrt-resistant haplotype in Gabon between 2011 and 2014 (>70%), even though chloroquine was no longer used. Despite a great diversity and variable distribution of the alleles linked to SP susceptibility, no resistant quintuple mutation haplotypes associated with IPTp-SP failure have been found in this period. Since ACT implementation, there is not yet artemisinin resistance in Southeast Gabon. However, the spread of antimalarial drug resistance from Southeast Asia to Africa, as described for chloroquine and SP, requires an intensive epidemiological survey.71,72
Acknowledgments
The authors thank the children and their parents who participated in the study and the staff of the pediatric wards of the health center laboratories of Lastourville, the regional hospitals, and Paul Moukambi and Amissa Bongo in Koulamoutou and Franceville, respectively. They are also grateful to the staff of the Unit of Evolution, Epidemiology and Parasitic Resistances at International Medical Research Center of Franceville (CIRMF), the Parasitology and Entomology Unit of Biomedical Research Institute of Army, and the National Malaria Reference Center of Marseille.
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
WHO: World Malaria Report 2016. Geneva: World Health Organization. 2016. | ||
Ashley EA, Dhorda M, Fairhurst RM, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2015;371(5):411–423. | ||
Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med . 2009;361(5):455–467. | ||
Sisowath C, Petersen I, Veiga MI, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. 2009;199(February 2008):750–757. | ||
Pradines B, Dormoi J, Briolant S, Bogreau H, Rogier C. La résistance aux antipaludiques. Rev Francoph des Lab. 2010;2010(422):51–62. | ||
Kublin JG, Dzinjalamala FK, Kamwendo DD, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185(3):380–388. | ||
Braun V, Rempis E, Schnack A, et al. Lack of effect of intermittent preventive treatment for malaria in pregnancy and intense drug resistance in western Uganda. Malar J. 2015;14(1):372. | ||
Gesase S, Gosling RD, Hashim R, et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in Northern Tanzania and the emergence of dhps resistance mutation at codon 581. PLoS One. 2009;4(2):e4569. | ||
Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):30–31. | ||
Ménard D, Khim N, Beghain J, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. 2016 ;374(25):2453–2464. | ||
Takala-harrison S, Jacob CG, Arze C, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. 2015;211(5):670–679. | ||
Taylor SM, Parobek CM, De Conti DK, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211(5):680–688. | ||
Torrentino-Madamet M, Fall B, Benoit N, et al. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012-2013. Malar J. 2014;13:472. | ||
Boussaroque A, Fall B, Madamet M, et al. Emergence of mutations in the K13 propeller gene of Plasmodium falciparum isolates from Dakar, Senegal in 2013-2014. Antimicrob Agents Chemother. 2015;60(1):624–627. | ||
Torrentino-madamet M, Collet L, Lepère F, Benoit N, Amalvict R, Ménard D. K13-Propeller polymorphisms in Plasmodium falciparum isolates from patients in Mayotte in 2013 and 2014. 2015;59(12):7878–7881. | ||
Ocan M, Bwanga F, Okeng A, et al. Prevalence of K13-propeller gene polymorphisms among Plasmodium falciparum parasites isolated from adult symptomatic patients in northern Uganda. BMC Infect Dis. 2016:16(1):428. | ||
Ouattara A, Kone A, Adams M, et al. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. 2015;92(6):1202–1206. | ||
Kamau E, Campino S, Amenga-Etego L, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-saharan Africa. J Infect Dis. 2015;211(8):1352–1355. | ||
Muwanguzi J, Henriques G, Sawa P, Bousema T, Sutherland CJ. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar J. 2016; 15:36. | ||
Hawkes M, Conroy AL, Opoka RO, et al. Slow clearance of Plasmodium falciparum in severe pediatric malaria, Uganda, 2011–2013 Michael. Emerg Infect Dis. 2015;21(7):1237–1239. | ||
Dorkenoo AM, Yehadji D, Agbo YM, et al. Therapeutic efficacy trial of artemisinin-based combination therapy for the treatment of uncomplicated malaria and investigation of mutations in k13 propeller domain in Togo, 2012–2013. Malar J. 2016:15331. | ||
Maïga-Ascofaré O, May J. Is the A578S single-nucleotide polymorphism in K13-propeller a marker of emerging resistance to artemisinin among Plasmodium falciparum in Africa? J Infect Dis. 2016;213:165–166. | ||
Li J, Chen J, Xie D, et al. Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int J Parasitol Drugs Drug Resist. 2016;6(1):54–59. | ||
Bouyou-Akotet MK, Mawili-Mboumba DP, de Dieu Tchantchou T, Kombila M. High prevalence of sulfadoxine/pyrimethamine-resistant alleles of Plasmodium falciparum isolates in pregnant women at the time of introduction of intermittent preventive treatment with sulfadoxine/pyrimethamine in Gabon. J Antimicrob Chemother. 2010;65(3):438–441. | ||
Lekana-Douki JB, Boutamba SDD, Zatra R, et al. Increased prevalence of the Plasmodium falciparum Pfmdr1 86N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether-lumefantrine and artesunate-mefloquine. Infect Genet Evol. 2011;11(2):512–517. | ||
Ramharter M, Schuster K, Bouyou-Akotet MK, et al. Malaria in pregnancy before and after the implementation of a national IPTp program in Gabon. Am J Trop Med Hyg. 2007;77(3):418–422. | ||
Frank M, Lehners N, Mayengue PI, et al. A thirteen-year analysis of Plasmodium falciparum populations reveals high conservation of the mutant pfcrt haplotype despite the withdrawal of chloroquine from national treatment guidelines in Gabon. Malar J. 2011; 10(1):304. | ||
Mawili-Mboumba DP, Ngomo JMN, Maboko F, et al. Pfcrt 76T and pfmdr1 86Y allele frequency in Plasmodium falciparum isolates and use of self-medication in a rural area of Gabon. Trans R Soc Trop Med Hyg. 2014;108(11):729–734. | ||
Mombo-Ngoma G, Oyakhirome S, Ord R, et al. High prevalence of dhfr triple mutant and correlation with high rates of sulphadoxine-pyrimethamine treatment failures in vivo in Gabonese children. Malar J. 2011;10(1):123. | ||
Ngomo JN, Mawili-mboumba DP, Bondoukwe NPM, et al. Increased prevalence of mutant allele Pfdhps 437G and Pfdhfr triple mutation in Plasmodium falciparum isolates from a rural area of Gabon, three years after the change of malaria treatment policy. 2016;2016:9694372. | ||
Planche T, Krishna S, Kombila M, et al. Comparison of methods for the rapid laboratory assessment of children with malaria. Am J Trop Med Hyg. 2001;65(5):599–602. | ||
Parola P, Pradines B, Simon F, et al. Antimalarial drug susceptibility and point mutations associated with drug resistance in 248 Plasmodium falciparum isolates imported from Comoros to Marseille, France in 2004–2006. Am J Trop Med. 2007;77(3):431–437. | ||
Boussaroque A, Fall B, Madamet M, et al. Prevalence of anti-malarial resistance genes in Dakar, Senegal from 2013 to 2014. Malar J. 2016;15:347. | ||
Tinto H, Bosco J, Erhart A, et al. Relationship between the Pfcrt T76 and the Pfmdr-1 Y86 mutations in Plasmodium falciparum and in vitro/in vivo chloroquine resistance in Burkina Faso, West Africa. Infect Genet Evol. 2003;3(4):287–292. | ||
Basco LK, Ringwald P. Molecular epidemiology of malaria in Cameroon. X . Evaluation of Pfmdr1 mutations as genetic markers for resistance to amino alcohols and artemisinin derivatives. 2002;66(6):667–671. | ||
Mawili-mboumba DP, Akotet MKB, Kendjo E, Nzamba J, Medang MO. Increase in malaria prevalence and age of at risk population in different areas of Gabon. Malar J. 2013;12:3. | ||
Maghendji Nzondo S, Kouna LC, Mourembou G, et al. Malaria in urban, semi-urban and rural areas of southern of Gabon : comparison of the Pfmdr 1 and Pfcrt genotypes from symptomatic children. Malar J. 2016;15(1):420. | ||
Maghendji-nzondo S, Nzoughe H, Lemamy GJ, et al. Prevalence of malaria, prevention measures, and main clinical features in febrile children admitted to the Franceville Regional. 2016;23:32. | ||
Ceesay SJ, Casals-pascual C, Erskine J, et al. Changes in malaria indices between 1999 and 2007 in The Gambia : a retrospective analysis. Lancet. 2007;372(9649):1545–1554. | ||
Hay SI, Guerra CA, Tatem AJ, Atkinson PM, Snow RW. Urbanization, malaria transmission and disease burden in Africa. Nat Rev Microbiol. 2005;3(1):81–90. | ||
Mott KE, Desjeux P, Moncayo A, Ranque P, de Raadt P. Parasitic diseases and urban development. Bull World Health Organ. 1990;68(6):691–698. | ||
Wilson ML, Krogstad DJ, Arinaitwe E, et al. Urban malaria: understanding its epidemiology, ecology, and transmission across seven diverse ICEMR network sites. Am J Trop Med Hyg. 2015;93(Suppl 3):110–123. | ||
Neiderud C-J. How urbanization affects the epidemiology of emerging infectious diseases. Infect Ecol Epidemiol. 2015;5:27060. | ||
Sondo P, Derra K, Tarnagda Z, et al. Dynamic of Plasmodium falciparum chloroquine resistance transporter gene Pfcrt K76T mutation five years after withdrawal of chloroquine in Burkina Faso. Pan Afr Med J. 2015;21:101. | ||
Lucchi NW, Komino F, Okoth SA, et al. In vitro and molecular surveillance for antimalarial drug resistance in Plasmodium falciparum parasites in western Kenya reveals sustained artemisinin sensitivity and increased chloroquine sensitivity. Antimicrob Agents Chemother. 2015;59(12):7540–7547. | ||
Tumwebaze P, Conrad MD, Walakira A, et al. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother. 2015;59(6):3018–3030. | ||
Zatra R, Lekana-douki JB, Lekoulou F, Bisvigou U, Ngoungou EB, Ndouo FST. In vitro antimalarial susceptibility and molecular markers of drug resistance in Franceville, Gabon. BMC Infect Dis. 2012;12(1):307. | ||
Sisowath C, Strömberg J, Mårtensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis. 2005;191(6):1014–1017. | ||
Fall B, Diawara S, Sow K, et al. Ex vivo susceptibility of Plasmodium falciparum isolates from Dakar, Senegal, to seven standard anti-malarial drugs. Malar J. 2011;10(1):310. | ||
Wurtz N, Fall B, Pascual A, et al. Prevalence of molecular markers of Plasmodium falciparum drug resistance in Dakar, Senegal. 2012:11(1):197. | ||
Fall B, Pascual A, Sarr FD, et al. Plasmodium falciparum susceptibility to anti-malarial drugs in Dakar, Senegal, in 2010 : an ex vivo and drug resistance molecular markers study. Malar J. 2012:107. | ||
Bouyou-Akotet MK, Tshibola ML, Mawili-Mboumba DP, et al. Frequencies of dhfr/dhps multiple mutations and Plasmodium falciparum submicroscopic gametocyte carriage in Gabonese pregnant women following IPTp-SP implementation. Acta Parasitol. 2015;60(2):218–225. | ||
Tsumori Y, Ndounga M, Sunahara T, et al. Plasmodium falciparum: differential selection of drug resistance alleles in contiguous urban and peri-urban areas of Brazzaville, republic of Congo. PLoS One. 2011;6(8):e23430. | ||
Nsimba B, Jafari-Guemouri S, Malonga DA, et al. Epidemiology of drug-resistant malaria in Republic of Congo: using molecular evidence for monitoring antimalarial drug resistance combined with assessment of antimalarial drug use. Trop Med Int Heal. 2005;10(10):1030–1037. | ||
Alker AP, Kazadi WM, Kutelemeni AK, Bloland PB, Tshefu K, Meshnick SR. dhfr and dhps genotype and sulfadoxine-pyrimethamine treatment failure in children with falciparum malaria in the Democratic Republic of the Congo. Trop Med Int Health. 2009;13(11):1384–1391. | ||
Tessema SK, Kassa M, Kebede A, Mohammed H. Declining trend of Plasmodium falciparum dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant alleles after the withdrawal of sulfadoxine-pyrimethamine in North Western Ethiopia. PLoS One. 2015;10(10):1–13:e0126943. | ||
Koukouikila-Koussounda F, Bakoua D, Fesser A, Nkombo M, Vouvoungui C, Ntoumi F. High prevalence of sulphadoxine-pyrimethamine resistance-associated mutations in Plasmodium falciparum field isolates from pregnant women in Brazzaville, Republic of Congo. Infect Genet Evol. 2015;33:32–36. | ||
Aubouy A, Jafari S, Huart V, et al. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine–pyrimethamine treatment efficacy. J Antimicrob Chemother. 2003;52(1):43–49. | ||
Gutman J, Kalilani L, Taylor S, et al. The A581 G mutation in the gene encoding Plasmodium falciparum dihydropteroate synthetase reduces the effectiveness of sulfadoxine-pyrimethamine preventive therapy in Malawian pregnant women. J Infect Dis. 2015;211(12):1997–2005. | ||
Vinayak S, Alam T, Mixson-Hayden T, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 2010;6(3):e100080. | ||
Artimovich E, Schneider K, Taylor TE, et al. Persistence of sulfadoxine-pyrimethamine resistance despite reduction of drug pressure in Malawi. J Infect Dis. 2015;212(5):694–701. | ||
Kaingona-Daniel EPS, Gomes LR, Gama BE, et al. Low-grade sulfadoxine–pyrimethamine resistance in Plasmodium falciparum parasites from Lubango, Angola. Malar J. 2016;15:309. | ||
Bouyou-Akotet MK, Mawili-Mboumba DP, Kendjo E, et al. Decrease of microscopic Plasmodium falciparum infection prevalence during pregnancy following IPTp-SP implementation in urban cities of Gabon. Trans R Soc Trop Med Hyg. 2016;110(6);333–342. | ||
Cooper RA, Conrad MD, Watson QD, et al. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother. 2015;59(8):5061–5064. | ||
Mohon AN, Alam MS, Bayih AG, et al. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009-2013). Malar J. 2014;13(1):431. | ||
Escobar C, Pateira S, Lobo E, et al. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One. 2015;10(3):2–7. | ||
Mbaye A, Dieye B, Ndiaye YD, et al. Selection of N86F184D1246 haplotype of Pfmrd1 gene by artemether – lumefantrine drug pressure on Plasmodium falciparum populations in Senegal. Malar J. 2016;15(1):433. | ||
Madamet M, Kounta MB, Wade KA, et al. Absence of association between polymorphisms in the K13 gene and the presence of Plasmodium falciparum parasites at day 3 after treatment with artemisinin derivatives in Senegal. Int J Antimicrob Agents. 2017;49(6):754–756. | ||
Plucinski MM, Talundzic E, Morton L, et al. Efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for treatment of uncomplicated malaria in children in Zaire and Ulge provinces, Angola. Antimicrob Agents Chemother. 2015;59(1):437–443. | ||
Sutherland CJ, Lansdell P, Sanders M, et al. Pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria given artemether-lumefantrine in the UK. Antimicrob Agents Chemother. 2017;61(3). pii:e02382. | ||
Mita T, Venkatesan M, Ohashi J, et al. Limited geographical origin and global spread of sulfadoxine-resistant dhps alleles in Plasmodium falciparum populations. J Infect Dis. 2011;204(12):1980–1988. | ||
Wootton JC, Feng X, Ferdig MT. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. 2002;418(6895):320–323. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

