Back to Journals » OncoTargets and Therapy » Volume 13
Prevalence of ESR1 Mutation in Chinese ER-Positive Breast Cancer
Authors Zhu W, Ren C, Wang Y, Wen L, Zhang G, Liao N
Received 7 October 2019
Accepted for publication 19 December 2019
Published 21 January 2020 Volume 2020:13 Pages 615—621
DOI https://doi.org/10.2147/OTT.S233662
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Wenzhen Zhu,* Chongyang Ren,* Yulei Wang, Lingzhu Wen, Guochun Zhang, Ning Liao
Department of Breast Cancer, Guangdong Provincial People’s Hospital & Guangdong Academy of Medical Sciences, Guangzhou, 510080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ning Liao
Department of Breast Cancer, Guangdong Provincial People’s Hospital & Guangdong Academy of Medical Sciences, 106 Zhongshan Er Road, Guangzhou 510080, People’s Republic of China
Email [email protected]
Background: ESR1 mutation and its possible relation to endocrine therapy resistance in ER-positive breast cancers have been studied with respect to genetic sequencing data from Western patients but rarely from Chinese patients. This study aimed to investigate the prevalence of ESR1 mutation in Chinese primary and metastatic ER-positive breast cancer.
Methods: Tumor samples from 297 primary breast cancer (PBC) patients and blood samples from 43 metastatic breast cancer (MBC) patients were obtained to perform whole exon sequencing of the ESR1 gene through next-generation sequencing (NGS). Clinicopathological features of MBC patients were listed and grouped to explore potential factors in ESR1 mutations.
Results: A total of 15 ESR1 variations, including 11 point mutations, 1 in-frame deletion mutation, 1 synonymous mutation, and 2 amplifications were identified in 13 patients. The ESR1 mutation rate was 1% (3/297) in PBC patients and 18.6% (8/43) in MBC patients. All ESR1 point mutations occurred in the estrogen receptor ligand-binding domain. Six (54.5%) of the 11 point mutations were hotspot mutations. Among all MBC patients, the ESR1 mutation rate in those who had a treatment history using aromatase inhibitors (AI) was significantly higher than those who did not (25.8% versus 0%, P=0.015). Moreover, the ESR1 mutation rate in those who received AI treatment over a period of 12 months was significantly higher than in those whose treatment lasted less than 12 months [36.3% versus 0%, P< 0.001].
Conclusion: ESR1 mutations were more frequently observed in the circulating cell-free DNA of MBC patients than in PBC patients among the Chinese cohort, and higher among those pretreated with AI, suggesting that such mutations may undergo selection during AI treatment.
Keywords: breast cancer, ESR1 mutation, endocrine therapy resistance, NGS, aromatase inhibitors
Introduction
Breast cancer accounted for 15.1% of all newly diagnosed cancers in the Chinese woman cohort from 2009 to 2011.1 Endocrine therapy targeting estrogen receptors (ER) can significantly lower the risk of relapse in the early stages of breast cancer as well as improve the survival outcomes for patients with advanced breast cancer.2,3 However, it is reported that a certain number of endocrine-resistant breast cancers could occur after endocrine therapy, resulting in cancer recurrence or metastasis.4,5 Endocrine therapy is widely used to treat ER-positive breast cancers in the Chinese patient cohort, although the ER-positive rate in Chinese breast cancer patients is lower than in Western patient (50–60% vs 70%).3,6 Nevertheless, this therapy is used while knowing little about endocrine therapy resistance in the Chinese patient cohort.
There are multiple reasons for endocrine therapy resistance, such as loss of ERα,7 up-regulation of ERβ8 and cross-talk between ER-genomic and growth factor pathways.9 As a ligand-activated nuclear hormone receptor, the ERα can regulate cell growth, thus affect survival and metastasis in most breast cancers.3 Many studies have found that variations of the ERα encoding gene ESR1 might be a critical factor leading to endocrine therapy resistance.10–12 Among different types of ESR1 genomic variations,13,14 point missense mutations in ERα ligand-binding domains (LBD) have been most commonly observed.15,16 ESR1 mutations may induce ligand-independent activation of ERα, which further leads to a conformational change of ER and possible endocrine therapy resistance of ER-positive breast cancers.3,11,17 However, those findings were mostly based upon Western patients. The ESR1 mutation situation in Chinese breast cancer patients remains unclear. Caution should be taken when using these ESR1 mutation data to assist treatment decisions concerning the utilization of endocrine therapy with Chinese breast cancer patients.
The identification of ESR1 mutation is affected by the genetic detection method. Although digital PCR is commonly used to identify known ESR1 mutations of circulating tumor DNA (ctDNA),17–19 the next-generation sequencing (NGS) assay can enable higher throughput and more comprehensive gene sequencing.20 NGS has been used to detect novel ESR1 mutations in ctDNA with a reported error rate as low as 0.01%.21–23 Therefore, the purpose of this study was to perform NGS assay on all exons of the ESR1 gene taken from primary tumor samples and blood samples of metastatic breast cancer to identify the prevalence of ESR1 mutation in Chinese breast cancer patients.
Materials and Methods
Patients and Samples
A total of 340 patients, including 297 primary breast cancer (PBC) patients and 43 metastatic breast cancer (MBC) patients, were recruited for this study. Tumor tissue samples were collected from PBC patients, while 10 mL peripheral blood samples were collected from MBC patients and stored in Struck tubes. Critical pathological characteristics, such as pathological grade and progesterone receptor (PR) status, were determined through standard pathological tests24 and shown in Table 1. Human epidermal growth factor receptor-2 (HER2) status was determined by immunohistochemistry (IHC) test. If an equivocal IHC result was found, fluorescence in situ hybridization (FISH) test was implemented to further determine HER2 status.25 Progression-free survival (PFS), defined as the time from starting treatment after ESR1 mutations to documented disease progression or death, was analyzed based on the data cutoff in April 2018. This study was conducted in accordance with the Declaration of Helsinki and approved by the ethical review board of the Guangdong Provincial People’s Hospital. All patients signed the informed consent before the study.
 |
Table 1 Pathological Characteristics of Patients Enrolled |
DNA Sample Extraction
For tumor tissue samples, the DNA sample was extracted using a QIAamp DNA FFPE tissue kit (Qiagen, California, US). DNA concentration was measured through a Qubit dsDNA assay (Life Technologies, California, US). Blood samples were first kept at room temperature for 2 hrs before DNA extraction. The supernatant was transferred into a 15-mL centrifuge tube and then centrifuged for 10 min at 16,000 rpm at 4°C. Circulated cell-free DNA (cfDNA) was recovered from 4 to 5 mL of plasma using a QIAamp Circulating Nucleic Acid kit (Qiagen, California, US). Quantification of cfDNA was performed using a Qubit 2.0 Fluorimeter (manufacturer) with a double-stranded (dsDNA) HS assay kit (Life Technologies, Carlsbad, CA). At least 50ng of cfDNA was required for NGS library construction.
NGS Library Preparation and Capture-Based Targeted DNA Sequencing
The extracted DNA sample was subjected to end repair, phosphorylation and adaptor ligation. DNA fragments sized 200–400 bp were selected by AM-Pure beads (Agencourt AMPure XP Kit), and then subjected to hybridization with capture probe baits, hybrid selection with magnetic beads and PCR amplification. A bio-analyzer high-sensitivity DNA assay was subsequently performed to assess the quality and size of the DNA fragments. Indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., US) with pair-end reads.
Sequence Data Analysis
Sequence data were mapped to the human genome (hg19) using a BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using a GATK 3.2, MuTect, and VarScan, respectively. A plasma sample was compared against its own white blood cell control sample to identify somatic variants. Variants were filtered using the VarScan fpfilter pipeline, with loci having depths less than 100 filtered out. At least 2 and 5 supporting reads were needed for insertions and deletions (InDels) in plasma and tissue samples, respectively, while 8 supporting reads were needed for SNVs to be called in both plasma and tissue samples. According to the ExAC, 1000 Genomes, dbSNP, ESP6500SI-V2 database, variants with a population frequency over 0.1% were grouped as Single Nucleotide Polymorphisms (SNP) and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3.
The “normalized” ctDNA abundance was calculated using two different methods. First, the maximum allele frequency of all somatic mutations detected in each plasma sample was used as the surrogate of the proportion of ctDNA among all cfDNA extracted, and the total ctDNA amount was imputed by multiplying the total ctDNA amount by that proportion. Second, for mutations that were detected in both tissue and plasma samples, the ratio of allele frequencies found in plasma to that found in tissue was calculated, and the relative ctDNA abundance was defined as the average allele frequency ratio within each patient, capped at 1.
Statistical Analysis
The ratio and mode of ESR1 variation were calculated and descriptively reported according to the NGS results. All MBC patients were grouped according to different pathological characteristics such as prior hormonal therapy, metastasis sites, and HER2 status. χ2 tests were carried out to determine whether there was any difference in ESR1 mutation rates among the subgroups. The two-sided P values were nominal without adjustment for multiple testing. All analyses were hypothesis-driven. The significance level was set as 0.05. The clinical characteristics of ESR1-mutant MBC patients were reported. Statistical analysis was performed using SPSS statistical software (SPSS 13.0; SPSS Inc., Chicago, IL, USA).
Results
According to the results of the NGS assay, a total of 15 ESR1 variations in 13 patients were identified, including 13 ESR1 point mutations (substitutions or in-deletions) and 2 ESR1 amplifications. Among the 297 PBC patients, 5 ESR1 variant patients including 3 ESR1 point mutant patients and 2 CNV patients were identified, resulting in a 1% (3/297) ESR1 mutation rate among the PBC patients. 8 ESR1 mutation patients were identified among the 43 MBC patients, resulting in an 18.6% (8/43) ESR1 mutation rate in the MBC patients.
The 3 ESR1 mutations in the PBC patients comprised 3 mutation types: R555C, P535S, and A571 synonymous. The 10 ESR1 mutations in the 8 MBC patients comprised 6 mutations type: D538G (3/10), V422 in-frame deletion (1/10), E380Q (2/10), Y537N (1/10), Y537S (2/10), and L536H (1/10). All 13 ESR1 mutations were located within ER LBD, and 6 were hotspot mutations. The ESR1 mutations D538G, E380Q, and Y537S were recurrent mutations, which were all in the MBC patients. In addition, 2 MBC patients exhibited more than one ESR1 mutation (D538G plus V422 del, E380Q plus Y537N). Finally, for 3 ESR1-mutant MBC patients who could be matched to their pretreatment breast tumor tissues, no ESR1 mutation was detected in the earlier samples.
The differences between ESR1 mutation rates of subgroups of clinicopathological characteristics are shown in Table 2. ESR1 mutations were found to be significantly affected by AI treatment. The ESR1 mutation rate was significantly higher in MBC patients who had a history of AI treatment than in those who did not (P=0.015). In addition, MBC patients who had AI treatment for over 12 months had significantly higher ESR1 mutation rates than those whose treatment was less than 12 months (P<0.001). No significant difference in ESR1 mutation rates was observed between different HER2 status, menstrual status, metastasis sites, and previous endocrine exposure.
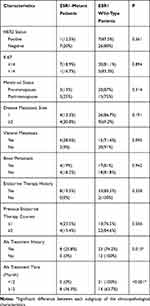 |
Table 2 Difference of ESR1 Mutation Rate in Metastatic Breast Cancer Patients Grouped by Pathological Characteristics |
The endocrine therapy histories for the 8 ESR1-mutant patients were retrospectively reviewed (Table 3). The 8 ESR1-mutant patients had an average AI treatment period of 25.2 months. The therapies of 5 patients were subsequently changed to stimulating estrogen receptor degradation drugs (SERDs) such as fulvestrant (Figures S1 1–8). All of them exhibited long survival time with stable disease. The other 3 patients died without further systemic therapy, with the longest survival time of 4 months (Figures S1 1–8).
 |
Table 3 Clinical Features of ER-Positive ESR1-Mutant Metastatic Patients (N=8) |
Discussion
Although ESR1 mutations have long been perceived to play a role in the endocrine therapy resistance of ER-positive breast cancers,13 ESR1 mutations in Chinese patients have been rarely studied. This is the first study investigating the prevalence of ESR1 mutation in ER-positive breast cancers in Chinese cohorts. In the present study, whole exon sequencing of ESR1 with NGS was performed on tumors or blood samples of 340 ER-positive PBC and MBC patients. ESR1 mutations were detected in 3.2% (11/340) of all enrolled patients. Specially, the ESR1 mutation rate was only 1% in the PBC patients, which was consistent with previous studies reporting that ESR1 mutations were seldom detected in Western PBC patients.10,12,15,16 This rate in the present study was slightly lower than the reported ER LBD mutation of 3% in the primary tumors (tumor without exposure to hormonal therapy) in the BOLERO-2 clinical trial.11 However, Jeselsohn and co-workers found no ER LBD mutation among 58 primary tumors.13 In this present study, the ESR1 mutation rate increased to 18.6% in the MBC patients, which was within the rate of 14–54% reported by previous studies based on Western MBC patients.11,13,15 Moreover, 3 ESR1-mutant MBC patients could be matched to their pretreatment breast tumor tissues where no ESR1 mutation was detected. The higher ESR1 mutation rate in the MBC patients indicates that ESR1 mutation might be associated with breast cancer progression.12,15,16 According to previous studies, the majority of ESR1 mutations were located within ER LBD, particularly hotspot mutations in residues Y537 and D538.26,27 In our study, all 13 ESR1 mutations were located within ER LBD, including 6 hotspot mutations. In addition, we detected two ESR1 amplifications in PBC patients but none in MBC patients, with a very low ESR1 amplification rate. This is close to the previously reported ESR1 amplification rate of 1.5–6% in breast cancer patients.15,28,29 A higher frequency of ESR1 amplification has been reported in PBC patients, which may be influenced by the detection method and scoring system.30
In previous studies, most patients received AI treatment prior to the detection of ESR1 mutation,10,12,15,16 which makes it necessary to explore the possible relation between the possible relation between ESR1 mutation and AI treatment. In the present study, ESR1 mutations were found to be associated with AI treatment. An ESR1 mutation rate of 25.8% was identified in the MBC patients who had a history of AI treatment, which was close to the average rate of 23% reported in previous studies.11,12,15,31 Meanwhile, the ESR1 mutation rate was 0% in MBC patients without a history of AI treatment. In addition, the ESR1 mutation rate was higher in MBC patients who had AI treatment for over 12 months than in those whose treatment was shorter than 12 months. The possible mechanism behind this may be that ESR1-mutant clones become prominent and harbor a growth advantage over other cells under the estrogen-deprivation caused by successive lines of AI treatment.17,19 Previous studies showed that ESR1 mutations were less frequently observed in patients who received AI treatment only at an adjuvant setting, suggesting the clonal selection of ESR1-mutant cancer cells under the estrogen-deprived condition in the metastatic tumors, but not in the small number of tumors during adjuvant treatment.17,19,23
A previous study reported that AI resistance might be associated with ESR1 mutation, and therefore ESR1-mutant patients should avoid subsequent AI treatment.17 Another study also found that the overall survival time of ESR1 mutation patients (i.e., D538G, 25.99 months; Y537S, 19.98 months; both D538G and Y537S mutations, 15.15 months) was shorter than ESR1 wild-type patients (32.1 months).19 For the 8 ESR1-mutant MBC patients in this study, the therapies of 5 patients were changed to SERDs such as fulvestrant. They exhibited long survival time with stable disease, indicating substantial benefits from this therapy change.
Several limitations in this study should be clarified. The present study had a relatively smaller sample size, which could be one of the reasons why several ESR1 mutation types (i.e., K303R and S463P) were not detected. Moreover, it prevented us from determining whether ESR1 mutation detection could predict sensitivity to specific hormone therapies or combinations, such as fulvestrant and/or CDK4/6 inhibitors. More studies should be performed to address whether there are clinical differences between ESR1 mutations, and to assess the lead time between the emergence of ESR1 mutations and the clinical progression of the disease through longitudinal sampling. In this study, we performed liquid biopsy rather than tumor biopsy in the MBC patient, in order to minimize the effect of inter-tumor heterogeneity on the accuracy of ESR1 mutation examination.32 To address the potential effect of sample selection, further study should focus on studying the concordance of ESR1 mutation pattern in MBC patients between different biopsies.
Conclusions
As the first study using NGS to identify ESR1 mutations in ER-positive breast cancer based on Chinese cohorts, our study showed that ESR1 mutations are rare in untreated primary tumors but are more likely to occur in metastatic patients with a history of AI treatment, indicating that such mutations may undergo selection during AI treatment. Future studies should identify molecular mechanisms of ESR1 mutations in AI resistance during the progression of ER-positive breast cancer.
Funding
This work was supported by funding from the National Natural Science Foundation of China (81602645; 81071851; 81001189), Natural Science Foundation of Guangdong Province (2016A030313768; 2018A030313292) and Research Funds from Guangzhou Municipal Science and Technology Project (201707010418; 201804010430).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348(24):2431–2442. doi:10.1056/NEJMra023246
3. Fanning SW, Mayne CG, Dharmarajan V, et al. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife. 2016;5. doi:10.7554/eLife.12792
4. Early Breast Cancer Trialists’ Collaborative G. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi:10.1016/S0140-6736(05)66544-0
5. Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi:10.1038/nrc2713
6. Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–e289. doi:10.1016/S1470-2045(13)70567-9
7. Lippman ME, Allegra JC. Quantitative estrogen receptor analyses: the response to endocrine and cytotoxic chemotherapy in human breast cancer and the disease-free interval. Cancer. 1980;46(12 Suppl):2829–2834.
8. Speirs V, Parkes AT, Kerin MJ, et al. Coexpression of estrogen receptor alpha and beta: poor prognostic factors in human breast cancer? Cancer Res. 1999;59(3):525–528.
9. Osborne CK, Schiff R. Growth factor receptor cross-talk with estrogen receptor as a mechanism for tamoxifen resistance in breast cancer. Breast. 2003;12(6):362–367. doi:10.1016/S0960-9776(03)00137-1
10. Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–1130. doi:10.1016/j.celrep.2013.08.022
11. Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi:10.1038/ng.2822
12. Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. doi:10.1038/ng.2823
13. Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12(10):573–583. doi:10.1038/nrclinonc.2015.117
14. Einarsdottir K, Darabi H, Li Y, et al. ESR1 and EGF genetic variation in relation to breast cancer risk and survival. Breast Cancer Res. 2008;10(1):R15. doi:10.1186/bcr1861
15. Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20(7):1757–1767. doi:10.1158/1078-0432.CCR-13-2332
16. Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-alpha: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73(23):6856–6864. doi:10.1158/0008-5472.CAN-13-1197
17. Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi:10.1126/scitranslmed.aac7551
18. Wang P, Bahreini A, Gyanchandani R, et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin Cancer Res. 2016;22(5):1130–1137. doi:10.1158/1078-0432.CCR-15-1534
19. Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016;2(10):1310–1315. doi:10.1001/jamaoncol.2016.1279
20. Kukita Y, Matoba R, Uchida J, et al. High-fidelity target sequencing of individual molecules identified using barcode sequences: de novo detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. DNA Res. 2015;22(4):269–277. doi:10.1093/dnares/dsv010
21. Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11(5):759–769. doi:10.1111/j.1755-0998.2011.03024.x
22. Toy W, Weir H, Razavi P, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7(3):277–287. doi:10.1158/2159-8290.CD-15-1523
23. Yanagawa T, Kagara N, Miyake T, et al. Detection of ESR1 mutations in plasma and tumors from metastatic breast cancer patients using next-generation sequencing. Breast Cancer Res Treat. 2017;163(2):231–240. doi:10.1007/s10549-017-4190-z
24. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi:10.1200/JCO.2009.25.6529
25. Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi:10.1200/JCO.2013.50.9984
26. Kumar S, Lindsay D, Chen QB, et al. Tracking plasma DNA mutation dynamics in estrogen receptor positive metastatic breast cancer with dPCR-SEQ. NPJ Breast Cancer. 2018;4:39. doi:10.1038/s41523-018-0093-3
27. Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Comparison of ESR1 mutations in tumor tissue and matched plasma samples from metastatic breast cancer patients. Transl Oncol. 2017;10(5):766–771. doi:10.1016/j.tranon.2017.07.004
28. Koboldt DC, Fulton RS, McLellan MD, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
29. Adelaide J, Finetti P, Charafe-Jauffret E, et al. Absence of ESR1 amplification in a series of breast cancers. Int J Cancer. 2008;123(12):2970–2972. doi:10.1002/ijc.v123:12
30. Holst F, Stahl PR, Ruiz C, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39(5):655–660. doi:10.1038/ng2006
31. Niu J, Andres G, Kramer K, et al. Incidence and clinical significance of ESR1 mutations in heavily pretreated metastatic breast cancer patients. Onco Targets Ther. 2015;8:3323–3328. doi:10.2147/OTT.S92443
32. De Mattos-arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–1735. doi:10.1093/annonc/mdu239
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
