Back to Journals » Breast Cancer: Targets and Therapy » Volume 8
Prevalence of CYP2D6*2, CYP2D6*4, CYP2D6*10, and CYP3A5*3 in Thai breast cancer patients undergoing tamoxifen treatment
Authors Charoenchokthavee W , Panomvana D, Sriuranpong V , Areepium N
Received 1 February 2016
Accepted for publication 21 March 2016
Published 8 August 2016 Volume 2016:8 Pages 149—155
DOI https://doi.org/10.2147/BCTT.S105563
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Pranela Rameshwar
Wanaporn Charoenchokthavee,1 Duangchit Panomvana,1 Virote Sriuranpong,2 Nutthada Areepium1
1Department of Pharmacy Practice, Faculty of Pharmaceutical Science, 2Medical Oncology Unit, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
Background: Tamoxifen (TAM) is used in breast cancer treatment, but interindividual variabilities in TAM-metabolizing enzymes exist and have been linked to single nucleotide polymorphisms in the respective encoding genes. The different alleles and genotypes of these genes have been presented for Caucasians and Asians. This study aimed to explore the prevalence of the incomplete functional alleles and genotypes of the CYP2D6 and CYP3A5 genes in Thai breast cancer patients undergoing TAM treatment.
Patients and methods: In total, 134 Thai breast cancer patients were randomly invited to join the Thai Tamoxifen Project. Their blood samples were collected and extracted for individual DNA. The alleles and genotypes were determined by real-time polymerase chain reaction with TaqMan® Drug Metabolism Genotyping Assays.
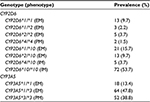
Results: The patients were aged from 27.0 years to 82.0 years with a body mass index range from 15.4 to 40.0, with the majority (103/134) in the early stage (stages 0–II) of breast cancer. The median duration of TAM administration was 17.2 months (interquartile range 16.1 months). Most (53%) of the patients were premenopausal with an estrogen receptor (ER) and progesterone receptor (PR) status of ER+/PR+ (71.7%), ER+/PR- (26.9%), ER-/PR+ (0.7%), and ER-/PR- (0.7%). The allele frequencies of CYP2D6*1, CYP2D6*2, CYP2D6*4, CYP2D6*10, CYP3A5*1, and CYP3A5*3 were 72.9%, 3.2%, 1.1%, 22.8%, 37.3%, and 62.7%, respectively, while the genotype frequencies of CYP2D6*1/*1, CYP2D6*1/*2, CYP2D6*2/*2, CYP2D6*4/*4, CYP2D6*1/*10, CYP2D6*2/*10, CYP2D6*4/*10, CYP2D6*10/*10, CYP3A5*1/*1, CYP3A5*1/*3, and CYP3A5*3/*3 were 9.7%, 2.2%, 3.7%, 1.5%, 15.7%, 9.7%, 3.7%, 53.7%, 13.4%, 47.8%, and 38.8%, respectively.
Conclusion: The majority (97.8%) of Thai breast cancer patients undergoing TAM treatment carry at least one incomplete functional allele, including 20.9% of the patients who carry only incomplete functional alleles for both the CYP2D6 and CYP3A5 genes. This research indicates the high prevalence of these defective alleles that are involved in TAM-metabolic pathways that might further affect TAM treatment.
Keywords: CYP450, SNPs, antihormone, oncology, pharmacogenetics, pharmacogenomics
Introduction
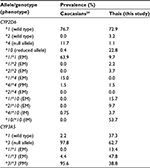
Tamoxifen (TAM) is a selective receptor modulator and has been used in hormone therapy for the prophylaxis and adjuvant and neoadjuvant treatment in breast cancer. It expresses a potent estrogenic antagonist effect on the estrogen receptor (ER) by competing with estrogen for binding to the ER and preventing cell growth1 in ER-positive breast cancer patients. Despite the fact that TAM provides significant clinical benefit, interindividual variability in its metabolism and effectiveness has been linked to single nucleotide polymorphisms (SNPs) in the genes encoding cytochrome P450 (CYP450) enzymes that are involved in TAM-metabolic pathways.2–5 CYP2D6 and CYP3A5 are the main CYP450 enzymes involved in the transformation of TAM from its conventional prodrug form to the active metabolites endoxifen (END) and 4-hydroxy-tamoxifen (4OHT) (Figure 1).1,6,7 Several studies have investigated the associations between polymorphisms in these two enzyme encoded genes and the clinical outcomes of breast cancer3,5,8–15 or the concentration of TAM and its metabolites (Figure 2).14,16–19 However, routine pharmacogenetic testing has not been suggested due to the inconclusive results obtained.20,21 These variations may result from differences in genotyping comprehensiveness, the DNA source, and the sample size. Nevertheless, previous studies revealed that the incomplete functional CYP2D6*4 and CYP3A5*3 alleles showed completely reduced enzyme activity, while CYP2D6*10 decreased its encoded enzyme function.22,23 The prevalence of these defective alleles was found to differ among racial populations. Thus, CYP2D6*4 was more prevalent in Caucasians than in Asians or Thais and CYP2D6*10 was more frequent in Asians (Table 1).8,14,16,24 CYP3A5*3 was the major allele in Caucasians and Asians, while CYP3A5*1 was the major allele in Thai populations (Table 1).9,14,24 Thai patients with CYP2D6*10/*10 had a lower END level than those patients with CYP2D6*1/*10 and CYP2D6*1/*1 (P=0.045)16 and had a shorter disease-free survival than heterozygous CYP2D6*10 or other genotypes (P=0.046).8 Previous studies have revealed the relationships between these gene polymorphisms and the clinical outcomes in Thai breast cancer patients.8,9,16 These results emphasize the opportunity to improve the pharmacological effectiveness of TAM, which is the most prescribed antihormone therapy for breast cancer treatment in Thailand. However, these studies were limited by their small sample sizes. The main objective of the present study was to determine the prevalence of the CYP2D6*2, CYP2D6*4, CYP2D6*10, and CYP3A5*3 defective alleles in a larger sample size of Thai breast cancer patients undergoing TAM treatment.
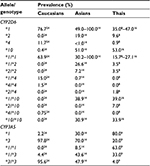  | Table 1 Prevalence of CYP2D6 and CYP3A5 polymorphisms in Caucasians, Asians, and Thais Note: Adapted from Copyright © 2013. PLoS One. Fernández-Santander A, Gaibar M, Novillo A, et al. Relationship between genotypes SULT1A2 and CYP2D6 and tamoxifen metabolism in breast cancer patients. 2013;8(7):e70183.24 © 2011 The Authors. British Journal of Clinical Pharmacology © 2011. The British Pharmacological Society. Lim JS, Chen XA, Singh O, et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. 2011;71(5):737–750.14 |
Patients and methods
Patient recruitment and sample preparation
Thai breast cancer patients who were taking TAM at 20 mg once daily and who visited the oncology outpatient clinic at King Chulalongkorn Memorial Hospital between February and March 2015 were randomly invited to join the Thai Tamoxifen Project. All patients were at least 18 years old with normal hepatic and renal functions (upper normal limit of aspartate aminotransferase and alanine aminotransferase ≤2, serum creatinine ≤1.2 mg/dL) in the 4 weeks prior to recruitment. Patients who were diagnosed for psychiatric illness or cognitive impairment were excluded from the study. For the remaining 134 patients who formed this study group, 10 mL of whole blood was drawn from each patient by a professional nurse and stored in a Vacutainer® K2EDTA tube (BD, Franklin Lakes, NJ, USA). DNA extraction was prepared from the buffy coat using a QIAamp® DNA Mini Kit (Qiagen NV, Venlo, the Netherlands). All DNA samples were stored at -20°C until analysis. The study protocol was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (IRB No 406/57). All patients provided written informed consent.
Genotype, phenotype, and data analysis
The genotype analysis was performed by real-time polymerase chain reaction (PCR) with TaqMan® Universal PCR Master Mix and TaqMan® Drug Metabolism Genotyping Assay Mix (CYP2D6*2, CYP2D6*10 and CYP3A5*3) with StepOnePlus® real-time PCR and Vii7 software (Thermo Fisher Scientific, Waltham, MA, USA). The PCR thermal cycling was performed as follows: 60°C for 30 seconds and 95°C for 10 minutes, and then, 40 cycles of 95°C for 15 seconds and 60°C for 1 minute before a final 60°C for 30 seconds. The rs numbers of CYP2D6*2, CYP2D6*10, and CYP3A5*3 were rs16947 (2850C>T), rs1065852 (100C>T), and rs776746 (6986A>G), respectively. CYP2D6*4 was determined by considering the haplotype of CYP2D6*2 (2850C>T) with CYP2D6*10 (100C>T). Phenotypes were classified as extensive metabolizer (EM) for normal enzyme activity and intermediate metabolizer (IM) and poor metabolizer (PM) for decreased and completely reduced enzyme activities, respectively (Table 2).22 Data analysis was conducted by descriptive statistics using the SPSS software (Version 22; IBM Corporation, Armonk, NY, USA).
Results
A total of 134 Thai breast cancer women undergoing TAM treatment formed the study group. Most of them were in the early stage of breast cancer (32.8% and 43.3% in stages I and II, respectively). Their mean age, weight, and BMI were 51.6±11.6 years (range 27.0–82.0 years), 60.0±10.7 kg (range 36.0–107.5 kg), and 24.0±4.3 (range 15.4–40.0), respectively. Their ER and progesterone receptor (PR) statuses were ER+/PR+ (71.6%), ER+/PR- (26.9%), ER-/PR+ (0.75%), and ER-/PR- (0.75%). The proportion of premenopausal patients (53%) was higher than the postmenopausal (36.6%) ones in this study group. Some patients had been treated with other breast cancer treatments before they received TAM, while nine patients (6.7%) had used TAM as monotherapy for breast cancer. The median duration of receiving TAM treatment was 17.2 months (interquartile range 16.1 months; range 0.8–62.1 months; Table 3). Of these 134 patients, the allelic frequencies of CYP2D6*1, CYP2D6*2, CYP2D6*4, CYP2D6*10, CYP3A5*1, and CYP3A5*3 were 72.9%, 3.2%, 1.1%, 22.8%, 37.3%, and 62.7%, respectively. CYP2D6*10/*10 and CYP3A5*1/*3 were the most common genotypes found in this study at 53.7% and 47.8%, respectively (Table 4). In total, 131 patients (97.8%) had at least one defective allele (CYP2D6*4, CYP2D6*10, and CYP3A5*3) that included 28 patients (20.9%) who had only defective alleles for both the CYP2D6 and CYP3A5 genes (25 patients with CYP2D6*10/*10 and CYP3A5*3/*3, two patients with CYP2D6*4/*4 and CYP3A5*3/*3, and one patient with CYP2D6*4/*10 and CYP3A5*3/*3), which was potentially responsible for their low enzyme activities compared with those patients who were carrying at least one normal functional allele (CYP2D6*1, CYP2D6*2, and CYP3A5*1).
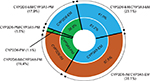
Moreover, the prevalence of the CYP2D6 phenotype was 41.0%, 57.5%, and 1.5% in the EM, IM, and PM enzyme activity categories, respectively, while the prevalence of the CYP3A5 phenotype was 61.2% and 38.8% for EM and PM, respectively. Note that CYP3A5-IM was not found in this study (Table 4). Furthermore, only 31 patients (23.1%) were categorized as EM for both the CYP2D6 and CYP3A5 genes, while the others (75.37%) were IM or PM for at least one gene, including the two patients (1.5%) who were PM for both the CYP2D6 and CYP3A5 genes (Table 5 and Figure 3).
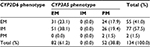  | Table 5 Phenotype frequency (%) of CYP2D6 with CYP3A5 (N=134) Abbreviations: EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer. |
Discussion
Prevalence of wild-type and mutant alleles including their genotypes was different depending on the racial origin of population (Table 1). However, the prevalence of these alleles was also different within the same population among studies. First, that the 9.7% frequency of the CYP2D6*1/*1 allele in Thai breast cancer patients found in this research is lower than the prevalence previously reported16 is probably the result of the simultaneous CYP2D6*2 determination. Previously, Areepium et al16 used only the CYP2D6*10 (100C>T) assay to discriminate the CYP2D6*10 SNP from the CYP2D6*1 wild type based on the quantitative PCR technique with the TaqMan® assay, while Chamnanphon et al8 used the AmpliChipTM CYP450 test that can detect several SNPs simultaneously, including CYP2D6*2 (2549delA), CYP2D6*4 (1846G>A), and CYP2D6*10 (100C>T).25 The prevalence of CYP2D6*1/*1 that was reported in these studies, therefore, depended on the number and position of SNPs that were included in each respective analytical platform, which varied among the three studies. Second, the higher prevalence of CYP2D6*10/*10 found in this study compared with previous studies in Thai breast cancer patients might be due to the larger number of patients who were recruited in this study. More patients helped identifying rare genotypes (CYP2D6*4/*4 and CYP2D6*4/*10) that were not reported from the previous study (Table 1).8 However, the summation of these genotype frequencies with fully functional alleles (CYP2D6*1/*1, CYP2D6*1/*2, and CYP2D6*2/*2) was 15.6%, which agreed with the previous studies.8,16 Determination of the CYP2D6*2 allele was useful for separating CYP2D6*2/*10 (EM), CYP2D6*4/*4 (PM), and CYP2D6*4/*10 (IM) from CYP2D6*1/*10 (EM), which was responsible for the low reported prevalence of CYP2D6*1/*10 (15.7%) in this study compared with the previous ones.8,16 The CYP2D6*4/*4 and CYP2D6*4/*10 alleles would potentially contribute to the low enzyme activities compared with CYP2D6*10/*10 allele and might differentially affect the concentration of TAM and its metabolites. There were seven patients whose genotypes were classified as CYP2D6*4 instead of CYP2D6*10 according to the simultaneous use of the CYP2D6*2 assay (Table 6). Gaedigk et al26 and Lyon et al25 suggested that the 100C>T SNP occurred on both the CYP2D6*4 and CYP2D6*10 alleles but they could be assigned during the absence of 1846G>A for the CYP2D6*4 allele.25,26
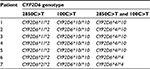  | Table 6 Determination of the CYP2D6*4 allele from the CYP2D6*2 and CYP2D6*10 assays |
The 1846G>A SNP was not used in this study due to the low prevalence of the CYP2D6*4 allele in Thai population, but the CYP2D6*4 allele was detected in this study by considering the haplotypes of 100C>T with 2850C>T,25 which were detected by the CYP2D6*10 and CYP2D6*2 assays, respectively. The CYP2D6*2 determination showed the following benefits in this study. First, the CYP2D6*2 assay helped to give a more accurate estimate of the prevalence of the CYP2D6*2 allele by discriminating between CYP2D6*2 and CYP2D6*1, the latter of which is another CYP2D6 wild type allele reported in Thai patients.8 Second, the CYP2D6*2 determination revealed the CYP2D6*2/*10, CYP2D6*4/*4, and CYP2D6*4/*10 genotypes, which have not been discovered by applying only the CYP2D6*10 assay. Third, the CYP2D6*4/*4 and CYP2D6*4/*10 alleles were found in this study, which potentially contributed to the different enzyme activities compared with CYP2D6*1/*10.
The prevalence of CYP2D6*10 was high in Thais, while the prevalence of CYP2D6*4 was high in Caucasians24 (Table 1), which subsequently raised CYP2D6*10/*10 to be the most common genotype in this study. This finding disagrees with the study in Caucasians that the CYP2D6*10/*10 allele has not been reported (Table 7).24 Moreover, the prevalence of the CYP3A5*1/*3 and CYP3A5*3/*3 alleles was also high in this study (47.8% and 38.8%, respectively) compared with CYP3A5*1/*1 (13.4%), which is in accordance with the previous studies in Caucasian24 and Asian14 populations (Tables 1 and 7) but disagrees with a previous report that the CYP3A5*1/*1 allele was the most common CYP3A5 genotype in Thai breast cancer patients.9 This disagreement might reflect the limited number of patients recruited in that study (30 patients) compared with the other studies.14,24 Furthermore, the low prevalence of the CYP3A5*1/*1 allele reported in Thai and Asian populations was due to the high prevalence of CYP3A5 polymorphisms in both Caucasians and Asians, including Thais. Moreover, it was found that 20.9% of these patients had only defective alleles, including the two patients who were PM for both CYP2D6 and CYP3A5 genes. Thus, Thai breast cancer patients undergoing TAM treatment potentially have a risk for a low activity of the enzymes involved in the TAM-metabolic pathway, especially for the CYP2D6 and CYP3A5 enzymes that are responsible for END formation.
  | Table 7 Prevalence of CYP2D6 and CYP3A5 polymorphisms in Caucasians and Thais Note: Adapted from Copyright © 2013. PLoS One. Fernández-Santander A, Gaibar M, Novillo A, et al. Relationship between genotypes SULT1A2 and CYP2D6 and tamoxifen metabolism in breast cancer patients. 2013;8(7):e70183.24 The phenotype classification in this table was defined by the authors of this article. Abbreviations: EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer. |
This study had some limitations. The frequency of the CYP2D6*4 allele was determined by considering the CYP2D6*2 (2850C>T) and CYP2D6*10 (100C>T) haplotypes without using the CYP2D6*4 (1846G>A) assay. According to the reported low prevalence of CYP2D6*4 in Thai breast cancer patients,8 these genotypes were classified as CYP2D6*2/*10. There were 13 patients (9.7%) who expressed CYP2D6*1/*2 (CYP2D6*2 assay) with CYP2D6*1/*10 (CYP2D6*10 assay) that were possibly carrying CYP2D6*2/*10 (2850C>T and 100C>T were located on different alleles) or CYP2D6*1/*4 (2850C>T and 100C>T were located on the same allele). These genotypes were classified as CYP2D6*2/*10 in this study due to the low prevalence of CYP2D6*4 in Thai patients.8 However, further investigations need to be performed to explore other possible allele patterns and confirm the genotype classification results of this study.
Conclusion
This research indicated a high prevalence of the CYP2D6 and CYP3A5 defective alleles in Thai breast cancer patients undergoing TAM treatment. Future studies should be focused on the concentration of TAM and its metabolites to reveal the true effect of the enzymes encoded by these gene polymorphisms on TAM treatment, which could be further applied for drug therapy monitoring in clinical settings.
Acknowledgments
This study was funded by the thesis grant for doctoral degree student of the National Research Council of Thailand (2015) and the 90th Anniversary of Chulalongkorn University Scholarship (2015).
Disclosure
The authors report no conflicts of interest in this work.
References
Klein DJ, Thorn CF, Desta Z, Flockhart DA, Altman RB, Klein TE. PharmGKB summary: tamoxifen pathway, pharmacokinetics. Pharmacogenet Genomics. 2013;23(11):643–647. | ||
Saladores PH, Precht JC, Schroth W, Brauch H, Schwab M. Impact of metabolizing enzymes on drug response of endocrine therapy in breast cancer. Expert Rev Mol Diagn. 2013;13(4):349–365. | ||
Rae JM, Drury S, Hayes DF, et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst. 2012;104(6):452–460. | ||
Karle J, Bolbrinker J, Vogl S, et al. Influence of CYP2D6-genotype on tamoxifen efficacy in advanced breast cancer. Breast Cancer Res Treat. 2013;139(2):553–560. | ||
Regan MM, Leyland-Jones B, Bouzyk M, et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1-98 trial. J Natl Cancer Inst. 2012;104(6):441–451. | ||
Westbrook K, Stearns V. Pharmacogenomics of breast cancer therapy: an update. Pharmacol Ther. 2013;139(1):1–11. | ||
Binkhorst L, Mathijssen RHJ, Ghobadi Moghaddam-Helmantel IM, et al. Quantification of tamoxifen and three of its phase-I metabolites in human plasma by liquid chromatography/triple-quadrupole mass spectrometry. J Pharm Biomed Anal. 2011;56(5):1016–1023. | ||
Chamnanphon M, Pechatanan K, Sirachainan E, et al. Association of CYP2D6 and CYP2C19 polymorphisms and disease-free survival of Thai post-menopausal breast cancer patients who received adjuvant tamoxifen. Pharmacogenomics Pers Med. 2013;6(1):37–48. | ||
Sensorn I, Sirachainan E, Chamnanphon M, et al. Association of CYP3A4/5, ABCB1 and ABCC2 polymorphisms and clinical outcomes of Thai breast cancer patients treated with tamoxifen. Pharmacogenomics Pers Med. 2013;6(1):93–98. | ||
Lum DWK, Perel P, Hingorani AD, Holmes MV. CYP2D6 genotype and tamoxifen response for breast cancer: a systematic review and meta-analysis. PLoS One. 2013;8(10):e76648. | ||
Province MA, Goetz MP, Brauch H, et al. CYP2D6 genotype and adjuvant tamoxifen: meta-analysis of heterogeneous study populations. Clin Pharmacol Ther. 2014;95(2):216–227. | ||
Madlensky L, Natarajan L, Tchu S, et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin Pharmacol Ther. 2011;89(5):718–725. | ||
Gjerde J, Geisler J, Lundgren S, et al. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer. 2010;10(1):313. | ||
Lim JS, Chen XA, Singh O, et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol. 2011;71(5):737–750. | ||
Teh LK, Mohamed NI, Salleh MZ, et al. The risk of recurrence in breast cancer patients treated with tamoxifen: polymorphisms of CYP2D6 and ABCB1. AAPS J. 2012;14(1):52–59. | ||
Areepium N, Panomvana D, Rungwanonchai P, Sathaporn S, Voravud N. Effects of CYP2D6 and UGT2B7 polymorphisms on pharmacokinetics of tamoxifen in Thai breast cancer patients. Breast Cancer (Dove Med Press). 2013;5:73–78. | ||
Zafra-Ceres M, de Haro T, Farez-Vidal E, et al. Influence of CYP2D6 polymorphisms on serum levels of tamoxifen metabolites in Spanish women with breast cancer. Int J Med Sci. 2013;10(7):932–937. | ||
Mürdter TE, Schroth W, Bacchus-Gerybadze L, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther. 2011;89(5):1–10. | ||
Barginear MF, Jaremko M, Peter I, et al. Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clin Pharmacol Ther. 2011;90(4):605–611. | ||
Ratain MJ, Nakamura Y, Cox NJ. CYP2D6 genotype and tamoxifen activity: understanding interstudy variability in methodological quality. Clin Pharmacol Ther. 2013;94(2):185–187. | ||
Hertz DL, McLeod HL, Irvin WJ Jr. Tamoxifen and CYP2D6: a contradiction of data. Oncologist. 2012;17(5):620–630. | ||
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. | ||
Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet. 2012;27(1):55–67. | ||
Fernández-Santander A, Gaibar M, Novillo A, et al. Relationship between genotypes SULT1A2 and CYP2D6 and tamoxifen metabolism in breast cancer patients. PLoS One. 2013;8(7):e70183. | ||
Lyon E, Gastier Foster J, Palomaki GE, et al. Laboratory testing of CYP2D6 alleles in relation to tamoxifen therapy. Genet Med. 2012;14(12):990–1000. | ||
Gaedigk A. Complexities of CYP2D6 gene analysis and interpretation. Int Rev Psychiatry. 2013;25(5):534–553. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.