Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 15
Prediction of Obstructive Lung Disease from Chest Radiographs via Deep Learning Trained on Pulmonary Function Data
Authors Schroeder JD , Bigolin Lanfredi R , Li T, Chan J, Vachet C , Paine R III, Srikumar V, Tasdizen T
Received 3 September 2020
Accepted for publication 11 December 2020
Published 5 January 2021 Volume 2020:15 Pages 3455—3466
DOI https://doi.org/10.2147/COPD.S279850
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Joyce D Schroeder,1 Ricardo Bigolin Lanfredi,2 Tao Li,3 Jessica Chan,1 Clement Vachet,4 Robert Paine III,5 Vivek Srikumar,3 Tolga Tasdizen2
1Department of Radiology and Imaging Sciences, School of Medicine, University of Utah, Salt Lake City, UT, USA; 2Department of Electrical and Computer Engineering, Scientific Computing and Imaging Institute (SCI), University of Utah, Salt Lake City, UT, USA; 3School of Computing, University of Utah, Salt Lake City, UT, USA; 4Biomedical Imaging and Data Analytics Core, SCI, University of Utah, Salt Lake City, UT, USA; 5Division of Pulmonary and Critical Care Medicine, School of Medicine, University of Utah, Salt Lake City, UT, USA
Correspondence: Joyce D Schroeder
Department of Radiology and Imaging Sciences, School of Medicine, University of Utah, 30 North 1900 East, Rm #1A71, Salt Lake City, UT 84132, USA
Tel +1 801 581 7553
Fax +1 801 581 2414
Email [email protected]
Background: Chronic obstructive pulmonary disease (COPD), the third leading cause of death worldwide, is often underdiagnosed.
Purpose: To develop machine learning methods to predict COPD using chest radiographs and a convolutional neural network (CNN) trained with near-concurrent pulmonary function test (PFT) data. Comparison is made to natural language processing (NLP) of the associated radiologist text reports.
Materials and Methods: This IRB-approved single-institution retrospective study uses 6749 two-view chest radiograph exams (2012– 2017, 4436 unique subjects, 54% female, 46% male), same-day associated radiologist text reports, and PFT exams acquired within 180 days. The Image Model (Resnet18 pre-trained with ImageNet CNN) is trained using frontal and lateral radiographs and PFTs with 10% of the subjects for validation and 19% for testing. The NLP Model is trained using radiologist text reports and PFTs. The primary metric of model comparison is the area under the receiver operating characteristic curve (AUC).
Results: The Image Model achieves an AUC of 0.814 for prediction of obstructive lung disease (FEV1/FVC < 0.7) from chest radiographs and performs better than the NLP Model (AUC 0.704, p< 0.001) from radiologist text reports where FEV1 = forced expiratory volume in 1 second and FVC = forced vital capacity. The Image Model performs better for prediction of severe or very severe COPD (FEV1 < 0.5) with an AUC of 0.837 versus the NLP model AUC of 0.770 (p< 0.001).
Conclusion: A CNN Image Model trained on physiologic lung function data (PFTs) can be applied to chest radiographs for quantitative prediction of obstructive lung disease with good accuracy.
Keywords: machine learning, chronic obstructive pulmonary disease, quantitative image analysis, natural language processing
Introduction
Deep learning techniques are rapidly being applied to medical image interpretation.1–3 Multiple studies show convolutional neural network (CNN) models developed to detect specific image features on chest radiographs.4–9 Subsequent evaluation of most models asks: “did the CNN model trained on radiologist-labels perform as well or better than the interpreting radiologist?” These approaches to supervised learning require costly and time-consuming “labeling” of disease by radiologists.
We pose the question:
can a CNN model based on physiologic measures of pulmonary disease be used to improve the identification of obstructive lung disease on chest radiograph images compared to the radiologist?
Chest radiographs are, in essence, a “missed screening opportunity” if COPD is present but not described by the radiologist.
COPD is the third leading cause of death worldwide but is often underdiagnosed.10–12 Airway inflammation, air trapping and emphysema may be secondary to smoking or environmental exposures and people with COPD are at increased risk of respiratory infections and cancer.13 Studies report a twofold to fourfold increase in lung cancer risk in patients with COPD compared to those without airflow obstruction.14 Lung cancer is the highest mortality cancer in the US and is often discovered at distant stage.15 The National Lung Screening Trial (NLST) showed a 20% reduction in lung cancer mortality for subjects imaged with CT compared to chest radiograph.16 However, although lung cancer screening with low-dose computed tomography (LDCT) is now recommended, few patients actually receive CT screening exams: for 2010–2015 fewer than 4% and in 2016 fewer than 2% of those eligible.17–19 COPD is typically diagnosed based on PFTs. However, these studies are performed in a minority of individuals at risk. Chest radiographs are the most common imaging study performed worldwide. Identification of individuals with COPD on chest radiographs alone would be a useful adjunct, and, offers an opportunity to target individuals for LDCT screening and/or smoking cessation programs.
Our hypothesis is that a deep learning algorithm trained using chest radiographs with annotation from PFTs (Image Model) will show greater accuracy for the prediction of COPD than text evaluation of the associated radiologist clinical reports (NLP Model, via both bidirectional recurrent neural networks and recent state-of-the-art transformer architecture models).20–22 The purpose is to develop a CNN Image model that can be used to augment radiology clinical reports with a quantitative prediction of obstructive lung disease, an important pulmonary disease associated with significant morbidity, mortality and increased risk of lung cancer.
Materials and Methods
Data Acquisition
This study is approved by the University of Utah Institutional Review Board, including a waiver of consent (the research and privacy risk of the research are no more than minimal), approval for the study plan for patient data confidentiality and compliance with the Declaration of Helsinki. This single-institution retrospective study (Figure 1) uses 6749 two-view chest radiographs, same-day associated radiologist text reports, and near-concurrent PFT exams for 4436 unique subjects. Due to insufficient numbers of subjects with post-bronchodilator PFTs, pre-bronchodilator PFTs are used to enrich the number of cases for training. Inclusion criteria are: pre-bronchodilator PFT exams from 2012 to 2017 with an electronic medical record (EMR) search showing a two-view chest radiograph within 180 days. Subjects with asthma or cystic fibrosis (n=200, <5%) are included.23 Lung transplant subjects are excluded.
For 70% of the study dataset, the PFT is within 15 days of the chest radiograph; other time differences are as follows: 1, 10, 20, 30, 60 and 90 days (51%, 66%, 72%, 79%, 87%, 92%, respectively). Table 1 shows the demographics of the 4436 unique study subjects, the first PFT occurring within the study timeline, and COPD diagnosis code in the EMR if present.
 |
Table 1 Demographics, Spirometry and EMR Diagnosis Code for N=4436 Unique Subjects (for Subjects with More Than One PFT Exam, Their First PFT Exam is Reported in This Table) |
PFTs
All PFTs in the study are pre-bronchodilator values, as used in multiple COPD studies.24–26 FEV1 is forced expiratory volume in 1 second. All ‘FEV1ʹ values reported in this paper are “percent-predicted” (%predFEV1) as provided by the institution’s PFT lab using the National Health and Nutrition Examination Survey (NHANESIII). FVC is forced vital capacity. COPD is defined as FEV1/FVC<0.7 and Global Initiative for Chronic Obstructive Lung Disease (GOLD) stages indicate the severity of disease based on the %predFEV1, ranging from GOLD stage I—mild to GOLD stage IV—very severe.27
Images
Chest radiograph exams in the initial dataset are filtered: 1) both posterior-anterior (PA) and lateral images must be present, 2) exams with greater than two images are excluded, and 3) only the pair of chest radiograph and PFT exams with the smallest date difference is used (maximum date 180 days).
Text
The associated complete radiologist text report, including indication, findings and impression, is acquired for the chest radiograph exams.
Dataset
Before filtering is applied, 10% of the subjects are selected for the validation set and 20% for the test set. There are 4436 unique subjects. After filtering, this results in 10% for validation and 19% for testing. The study uses a total of 6749 PFT – Image/Text pairs: 4773 pairs for training, 752 pairs for validation, and 1224 pairs for testing.
Data Preprocessing
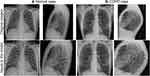
Images are cropped using the largest centralized square fitting within the image, resized to 256x256 pixels, and normalized using the mean and standard deviation of the ImageNet dataset.28 The dataset is augmented by horizontal flipping (PA images) and extracting random 224x224 crops. Figure 2 shows an example of original and preprocessed images. The text reports are converted into a sequence of tokens using the spaCy tokenizer (http://spacy.io/).
Model Implementation
Image Model
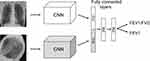
The Image model (Figure 3) uses PA and lateral chest radiographs as inputs to two independent CNNs based on the Resnet-18 model, with weights from a model pretrained on ImageNet for initialization with the final fully connected layer removed.29 The output layers of both CNNs are concatenated. The resulting vector is used as input to a block of fully connected layers with two hidden layers and softplus output non-linearity. The hidden layers use ReLU activation and dropout (with p=0.25).30 The outputs of the model are two positive real values, FEV1/FVC and %predFEV1. The overall model is trained end-to-end with an L1 loss. The batch size is 20, the initial learning rate is 0.0001, and the model is trained for 50 epochs using the Adam optimizer.31 The learning rate was reduced by a factor of 10 every time the loss plateaued for 5 epochs.
NLP Model
The NLP pipeline regresses raw text to the same two positive real values, FEV1/FVC and FEV1. We experiment with two strong NLP models: BiLSTM, which represents a standard design of recurrent neural networks, and RoBERTa, which represents recent state-of-the-art transformer architecture.20–22
With the BiLSTM model, we use the Common Crawl version of GloVe embeddings with 100 dimensions, combined with character-level embeddings convoluted by a 2-dimensional filter of size 5.32 The output character encodings of the convolutional network are 100 dimensional. The character encodings and token embeddings (GloVe) are combined by 2-layer highway networks, resulting in 200-dimensional token-level encodings.33 Such encodings are then processed by the BiLSTM with the same output dimensionality. The sequence of token encodings is aggregated via self-attention to form a representation of the entire text, which is fed into a single linear layer with a soft plus activation for the final regression output. For training, the model parameters and character-level embeddings were initialized randomly with uniform distribution (with gain=1).34 We train this model for 50 epochs with learning rate 0.0001 and dropout rate 0.1 using the Adam optimizer.35,36
With the RoBERTa model, we fine-tuned the pre-trained language model for this task. We used the base version of RoBERTa and take the encoding of the CLS token at the final layer as the text representation vector. This representation forms the input to a regression layer that has the same design as in the BiLSTM model. For training, we fine-tuned the model for 20 epochs with learning rate 0.000005, optimized by AdamW with a warm-up percent of 20%.37 The dropout rate for hidden states within the transformer model and before the last linear layer is 0.3.
During training, for both models, we choose the version with the best performance on the development set and report their results on the test set accordingly. We used grid search over the validation set to find the hyperparameters mentioned above.
Statistical Analysis
All models are trained with five different random seeds in order to assess the variability of results with the proposed approach. We compared the Image and NLP models using AUC (area under the receiver operating characteristic curve), accuracy and R2, with AUC as the primary metric. For FEV1/FVC, AUC and accuracy are measured by setting a threshold, FEV1/FVC <0.7, as a positive label for COPD diagnosis. For %predFEV1, AUC is measured by setting a threshold, FEV1 <0.5 (GOLD stage severe to very severe), only for cases that are classified as having COPD in the ground-truth. We compared the AUCs from the Image and NLP models using DeLong’s method for comparing two correlated ROC curves (pROC package in R version 3.6.1) with P < 0.05 considered a statistically significant difference. Bland–Altman plots are also provided.
Results
Mean age, sex ratios, and initial PFT values are similar between the training, validation and test datasets (Table 1). For the 4436 unique subjects, 66% show normal PFT values (2921) and 34% show obstructive lung disease by PFT values (GOLD stages I–IV total: 1515). The distribution of severity of disease by GOLD stages I–IV (mild to very severe) is 8%, 16%, 8% and 3%, respectively. Male/female ratios are similar for spirometry with normal PFT values and obstructive lung disease, the overall study ratio is 46% male/54% female. The mean age of subjects with normal PFTs (54.4) is lower than the mean age of subjects with disease severity GOLD stage I–III (64.0, 60.7, 60.1) but similar to subjects with GOLD stage IV (55.4).
Table 1 shows that less than half (49.5%) of subjects with PFTs indicating obstructive lung disease (GOLD stages I–IV total: 1515) had a diagnosis code of COPD in the EMR (GOLD stages I–IV total: 750). Diagnosis codes for COPD are present at higher levels for more severe disease: for GOLD stages I–IV the percentage of COPD diagnosis codes is 28.5%, 48.6%, 66.6% and 70.5%, respectively.
All reported results, including the scatter plots, are generated using the test set.
Performance of the Image Model
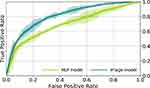
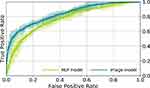
Figure 4 shows the receiver operating characteristic (ROC) curve for the trained Image Model using an FEV1/FVC threshold of 0.7 for ground-truth with COPD defined as FEV1/FVC <0.7. The average for five models trained with different random seeds is shown in the darker color line and one standard deviation of the results is shown in the lighter color band. The Image Model trained on PFT data results in good prediction of COPD from two-view chest radiograph exams (AUC 0.814±0.005) where AUC is area under the ROC curve. Accuracy for COPD is 0.749±0.008 and R2 FEV1/FVC is 0.415±0.016 (Table 2).
 |
Table 2 Results for Chosen Metrics for All Trained Models |
A confusion matrix for the Image Model in Table 3 shows the results of classification for different severities of disease (GOLD stages I–IV). The diagonal represents the line where all cases would be located if the model is perfect. The Image Model predicts normal PFTs for most actual normal cases (ground-truth from PFTs). There are 596.0±18.0 predicted normal cases with misclassifications mostly of mild to moderate COPD (GOLD stage I: 24.2±6.9 cases and GOLD stage II: 80.4±14.0 cases), few misclassifications of severe disease (GOLD stage III: 15.4±1.5) and no misclassifications of very severe disease (GOLD stage IV: 0±0.0). For mild to moderate COPD, most misclassifications result in the Image Model predicting normal instead of COPD: 55.0±4.3 cases for GOLD stage I and 80.2±11.4 for GOLD stage II. For severe and very severe COPD, most misclassifications by the Image Model still indicate COPD but one GOLD stage lower than the ground-truth stage. For actual GOLD stage III, the Image Model predicts 48.6±3.1 cases of GOLD stage II. For actual GOLD stage IV, the Image Model predicts 63.0±7.6 cases of GOLD stage III.
Figure 5 shows the ROC curve for the trained Image Model using a %predFEV1 threshold of 50% for ground-truth, only for cases classified as having COPD in the ground-truth. %predFEV1 <50% is GOLD stage III or IV (severe to very severe disease). This evaluates the Image model in the task of determining the severity of disease, assuming that we know the subject has COPD. The AUC is 0.837±0.003 for determination of severe disease with R2 FEV1 0.512±0.004 (Table 2).
Figures 6 and 7 show scatter plots and Bland–Altman plots of the results of the Image Model for both output variables: FEV1/FVC and %predFEV1. When evaluating the training dataset only, the model had an R2 FEV1/FVC of 0.922±0.004 and an R2 FEV1 of 0.937±0.007, highlighting a gap between testing scores and training scores. The choice of small CNN depth for the tested model is related to the pronounced overfitting of the model when using deeper architectures and validating on the validation dataset. More training data, in theory, could help in reducing the gap between training scores and validation/test scores and allow the use of deeper architectures.
 |
Figure 7 Bland–Altman plots showing the error of the trained models on the test set as a function of the average between ground-truth and model predication. The blue dashed lines represent 1.96 standard deviations from the average for the error data points, while the green line represents the average error. These graphs are plotted using the library pyCompare (jaketmp/pyCompare v1.5.1; http://doi.org/10.5281/zenodo.4001461). |
Performance of the NLP Model
Figure 4 shows the corresponding ROC curve for the trained NLP Model using an FEV1/FVC threshold of 0.7 for ground-truth with COPD defined as FEV1/FVC <0.7. The NLP Model trained on PFT data results in moderate prediction of COPD (AUC 0.704±0.010). Accuracy for COPD is 0.669±0.016 and R2 FEV1/FVC is 0.185±0.013 with less predictive metrics compared to the Image Model (Table 2).
The confusion matrix for the NLP Model (Table 3) shows greater numbers of misclassified cases by GOLD stage severity that are at least two GOLD stages different from the ground-truth compared to the Image Model.
The corresponding ROC curve for the NLP Model in determining severe to very severe COPD in subjects with known COPD is shown in Figure 5. The AUC %predFEV1 is 0.770±0.007 and R2%predFEV1 is 0.348±0.013. NLP Model metrics are less predictive for obstructive lung disease compared to the Imaging Model.
Scatter plots and Bland–Altman plots of the results of the NLP Model in Figures 6 and 7 show the predictions of the NLP model for both output variables: FEV1/FVC and %predFEV1.
One preliminary version of the NLP Model also evaluated common keywords related to COPD in radiologist text reports, by frequency including “emphysema”, “hyperinflation”, “COPD”, ‘flattened diaphragms’, and “vascular pruning”, with frequency order determined by radiologist chest radiograph text report search (Nuance mPower by Montage, 2018.) The keywords did not improve the model and are not a part of the final results.
Comparison of the Image Model and the NLP Model
Differences between Image Model and NLP Model AUCs are statistically significant, P < 0.001, for prediction of obstructive lung disease (FEV1/FVC <0.7) and severe to very severe GOLD stage III–IV disease (%predFEV1 <50%) using DeLong’s test for two correlated ROC curves. We also tested if the averages of the squared errors of the Image and NLP regression models are different, since regression is used directly instead of classification, with paired Wilcox test results P < 0.001 for both FEV1/FVC and FEV1.
Figures 6 and 7 show that both models, to different extents, predict values towards the mean when compared with the ground-truth in the extremes of the range of values.
Discussion
The results of this investigation strongly support our hypothesis that a deep learning (DL) algorithm trained using chest radiograph images with annotations from near-concurrent physiologic measures of lung function shows greater accuracy for the prediction of COPD than NLP evaluation of the associated radiologist clinical text reports. Implications of our findings include:
Physiologic measures of pulmonary function can be used to train CNNs for identification of obstructive lung disease on imaging. Labeling datasets with physiologic parameters avoids the time-consuming labeling and interpretation tasks by the radiologist. More importantly, it eliminates the inherent bias and limitations in these radiologist labels. For example, radiologist labels of “emphysema” used in other labeling studies may be ambiguous, describing features of lucent lungs, which may be secondary to either emphysema or air trapping from small airways disease. Physiologic parameters provide a gold standard, with less bias, and allow the algorithm to evaluate for imaging features that may not be assessed by the radiologist. In the case of COPD, pulmonary function tests, specifically the FEV1/FVC ratio, define the presence and severity of COPD in the appropriate clinical setting per the World Health Organization.27 For those with airflow obstruction, severity is determined by the %predFEV1. Radiographic imaging features of obstructive lung disease, including high lung volumes (hyperinflation), flattened hemi-diaphragms, increased retrosternal clear space, and upper lung predominant lucency and vascular pruning are low spatial resolution features that are not lost in the image downsizing required for inputs to the CNN (Figure 2).
An Image Model CNN for prediction of COPD can add information to augment the radiologist report. The Image Model demonstrates better prediction for COPD compared to the NLP Model from radiologist text reports, suggesting that the DL Image Model can be used to augment the radiologist text reports by generating a prediction of COPD, when present. There are multiple reasons why radiologist reports may not include text indicating COPD. Exams are ordered for other indications (eg, “rule out pneumonia”) and COPD features, particularly if not severe, may not be recognized or assessed. Chest radiographs are a high-volume modality with often narrow indication, eg, “trauma”, and clinical service demands may require short, very directed reports. It is also possible that radiologists may not recognize airflow obstruction in many instances.
Deep learning can be used to consider routine chest radiographs a “screening opportunity” for obstructive lung disease. Our results show that a DL Image Model, while not perfect, improves detection of obstructive lung disease compared to current practice. By generating a numeric metric of disease (predicted FEV1/FVC), the routine chest radiograph can be considered a screening opportunity for COPD. Results can be used to direct patients to medical care of COPD, lung cancer screening (for those meeting age and smoking history criteria) or smoking cessation programs.
Our observations on the performance of PFT annotation for the CNN Image Model compare favorably to other relevant studies using deep learning. Radiologist-labels for “emphysema” are used in these studies to develop networks with reported AUC values of 0.815, 0.829, and 0.926 for the detection of emphysema.4 Our findings of significant under-diagnosis of COPD, with less than 50% of the subjects with COPD by spirometry (PFTs) having a COPD diagnosis code in the EMR in our study population, are similar to other studies.10
The limitations of this study include the study size, single-institution cohort, and time difference between the chest radiographs/text reports and the PFT exam (up to 180 days). Our study uses pre-bronchodilator PFTs to enrich the number of cases for training which does not distinguish between obstructive lung disease from COPD (emphysema or bronchitis) and that from asthma. Furthermore, our FEV1/FVC <0.7 threshold does not exclude patients with “mixed” lung disease—those with both obstructive and restrictive lung disease, eg, COPD and pulmonary fibrosis. The selection of the maximum 180-day date difference was made to increase the number of cases in the study. Although this time difference introduces the possibility of an acute illness, eg acute exacerbation or pneumonia, that is present on one modality but not the other, we found slightly improved DL Image Model performance with 180-day date difference than with 2-day and 10-day date differences because of the added number of training cases (not shown). Large, diverse multi-institutional cohorts will be required to evaluate the generalizability of the CNN Image Model.
Conclusion
In summary, our study findings show improved prediction of COPD for a CNN Image Model trained with physiologic data (PFTs) compared to NLP Model evaluation of radiologist text reports trained with PFTs. This comparison of models, to our knowledge, is a novel approach for assessing the identification of obstructive lung disease on chest radiographs. Our study conclusions using deep learning to numerically predict airflow obstruction on routine chest radiographs support the goal of better detection of disease. Results suggest that a CNN model trained on physiologic lung function data can be used to augment the clinical radiologist report for improved identification of COPD, an under-diagnosed disease and risk factor for lung cancer.
Acknowledgments
We thank Mike Mitchell (Pulmonary Lab), Associate Professor Angela Presson (Division of Epidemiology), Matt Zabriskie (Radiology Research) and Mingyuan Zhang (Medical Informatics) at the University of Utah for their contributions. The support and resources from the Biomedical Image and Data Analytics Core, the Scientific Computing and Imaging Institute, and the Center for High Performance Computing at the University of Utah are gratefully acknowledged.
Disclosure
Dr Robert Paine III reports grants from NHLBI, COPD Foundation, and Department of Veterans Affairs; personal fees from Partner Therapeutics, outside the submitted work. The authors report no other conflicts of interest in this work.
References
1. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi:10.1016/j.media.2017.07.005
2. Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. doi:10.1016/j.media.2016.05.004
3. Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal. 2017;35:159–171. doi:10.1016/j.media.2016.05.009
4. Rajpurkar P, Irvin J, Zhu K, et al. Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. CoRR. 2017.
5. Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi:10.1148/radiol.2017162326
6. Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In:
7. Yao L, Poblenz E, Dagunts D, et al. Learning to diagnose from scratch by exploiting dependencies among labels. CoRR. 2017.
8. Li Z, Wang C, Han M, et al.. Thoracic disease identification and localization with limited supervision. In:
9. Seah JCY, Tang JSN, Kitchen A, et al. Chest radiographs in congestive heart failure: visualizing neural network learning. Radiology. 2019;290(2):514–522. doi:10.1148/radiol.2018180887
10. Martinez CH, Mannino DM, Jaimes FA, et al. Undiagnosed obstructive lung disease in the United States. AnnalsATS. 2015;12(12):1788–1795. doi:10.1513/AnnalsATS.201506-388OC
11. Martinez FJ, O’Connor GT. Screening, case-finding, and outcomes for adults with unrecognized COPD. JAMA. 2016;315(13):1333–1334. doi:10.1001/jama.2016.3274
12. Quaderi SA, Hurst JR. The unmet global burden of COPD. Glob Health Epidemiol Genom. 2018;3(ed4). doi:10.1017/gheg.2018.1
13. Chatila WM, Thomashow BM, Minai OA, et al. Comorbidities in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2008;5(4):549–555. doi:10.1513/pats.200709-148ET
14. Gonzalez J, Marin M, Sanchez-Salcedo P, Zulueta JJ. Lung cancer screening in patients with chronic obstructive pulmonary disease. Ann Transl Med. 2016;4(8):160. doi:10.21037/atm.2016.03.57
15. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
16. The National Lung Cancer Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi:10.1056/NEJMoa1102873
17. Moyer VA, LeFevre ML, Siu AL, et al. Screening for lung cancer: U.S. preventative services task force recommendation statement. Ann Intern Med. 2014;160(5):330–338. doi:10.7326/M13-2771
18. Jemal A, Fedewa SA. Lung cancer screening with low-dose computed tomography in the United States—2010 to 2015. JAMA Oncol. 2017;3(9):1278–1281. doi:10.1001/jamaoncol.2016.6416
19. Pham D, Bhandari S, Oechsli M, et al. Lung cancer screening rates: data from the lung cancer screening registry. J Clin Oncol. 2018;36(15_suppl):6504. doi:10.1200/JCO.2018.36.15_suppl.6504
20. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi:10.1162/neco.1997.9.8.1735
21. Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE Trans Signal Process. 1997;45(11):2673–2681. doi:10.1109/78.650093
22. Liu Y, Ott M, Goyal N, et al. RoBERTa: a Robustly Optimized BERT Pretraining Approach. arXiv. 2019.
23. Regan EA, Hokanson JE, Murphy JR, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2011;7(1):32–43. doi:10.3109/15412550903499522
24. Tseng H, Henry TS, Veeraraghavan S, et al. Pulmonary function tests for the radiologist. RadioGraphics. 2017;37(4):1037–1058. doi:10.1148/rg.2017160174
25. Tashkin DP, Wang H, Halpin D, et al. Comparison of the variability of the annual rates of change in FEV1 determined from serial measurements of the pre- versus post-bronchodilator FEV1 over 5 years in mild to moderate COPD: results of the lung health study. Respir Res. 2012;13(1):70. doi:10.1186/1465-9921-13-70
26. Mannino DM, Diaz-Guzman E, Buist S. Pre- and post-bronchodilator lung function as predictors of mortality in the lung health study. Respir Res. 2011;12(1):136. doi:10.1186/1465-9921-12-136
27. Rabe KF, Hurd S, Anzueto A, et al. Global initiative for chronic obstructive lung disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Resp Crit Care Med. 2007;176(6):532–555. doi:10.1164/rccm.200703-456SO
28. Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. doi:10.1007/s11263-015-0816-y
29. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In:
30. Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, Salakhutdinov RR. Improving neural networks by preventing co-adaptation of feature detectors. arXiv. 2012.
31. Kingma DP, Ba J. Adam: a method for stochastic optimization. Arxiv:1412.6980.
32. Pennington J, Socher R, Manning C. Glove: global vectors for word representation. In:
33. Srivastava RK, Greff K, Schmidhuber J. Highway networks. arXiv. 2015.
34. Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks.
35. Srivastava N, Hinton G, Krizhevshk A, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958.
36. Kingma DP, Ba JL. Adam: a method for stochastic optimization. International Conference on Learning Representations (ICLR) 2015. arXiv. 2017.
37. Loshchilov I, Hutter F. Decoupled weight decay regularization. International Conference on Learning Representations (ICLR) 2019. arXiv. 2019.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.