Back to Journals » Cancer Management and Research » Volume 10
Predicting the response to neoadjuvant therapy for early-stage breast cancer: tumor-, blood-, and imaging-related biomarkers
Authors Tan W , Yang M, Yang H, Zhou F, Shen W
Received 17 May 2018
Accepted for publication 11 July 2018
Published 9 October 2018 Volume 2018:10 Pages 4333—4347
DOI https://doi.org/10.2147/CMAR.S174435
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Wenyong Tan,1,2 Ming Yang,3 Hongli Yang,2 Fangbin Zhou,2 Weixi Shen1
1Department of Oncology, Shenzhen Hospital of Southern Medical University, Shenzhen, People’s Republic of China; 2Clinical Medical Research Center, The Second Clinical Medical College (Shenzhen People Hospital), Jinan University, Shenzhen, People’s Republic of China; 3Shenzhen Jingmai Medical Scientific and Technique Company, Shenzhen, People’s Republic of China
Abstract: Neoadjuvant therapy (NAT) has been used increasingly in patients with locally advanced or early-stage breast cancer. However, the accurate evaluation and prediction of response to NAT remain the great challenge. Biomarkers could prove useful to identify responders or nonresponders, or even to distinguish between early and delayed responses. These biomarkers could include markers from the tumor itself, such as versatile proteins, genes, and ribonucleic acids, various biological factors or peripheral blood cells, and clinical and pathological features. Possible predictive markers could also include multiple features from functional imaging, such as standard uptake values in positron emission tomography, apparent diffusion coefficient in magnetic resonance, or radiomics imaging biomarkers. In addition, cells that indirectly present the immune status of tumor cells and/or their host could also potentially be used as biomarkers, eg, tumor-infiltrating lymphocytes, tumor-associated macrophages, and myeloid-derived suppressor cells. Though numerous biomarkers have been widely investigated, only estrogen and/or progesterone receptors and human epidermal growth factor receptor have been proven to be reliable biomarkers to predict the response to NAT. They are the only biomarkers recommended in several international guidelines. The other aforementioned biomarkers warrant further validation studies. Some multigene profiling assays that are commercially available, eg, Oncotype DX and MammaPrint, should be used with caution when extrapolated to NAT settings. A panel of combined multilevel biomarkers might be able to predict the response to NAT more robustly than individual biomarkers. To establish such a panel and its prediction model, reliable methods and extensive clinical validation are warranted.
Keywords: breast cancer, drug therapy, biomarker, predictive factor
Introduction
Breast cancer accounts for ~30% of female malignant tumors both in USA1 and China.2 Treatment of breast cancer includes locoregional resection, with or without radiotherapy as well as systemic therapies such as chemotherapy, endocrine therapy, biological targeting agents, and a combination of the above. The need for the selection of local and systemic therapies depends mainly on various clinical, pathological, and molecular features. Markers are served as surrogates of these features for establishing prognostics and predicting outcomes.3 Prognostic factors may help select patients most likely to benefit from adjuvant therapy, while predictive factors can be valuable to predict the most efficacious therapy or measure response to therapy early in the course of treatment.4,5
Neoadjuvant therapy (NAT), which corresponds to the administration of systemic anticancer agents prior to local treatment, has been recommended as a general approach in locally advanced-stage diseases.6 Though NAT of breast cancer has been shown to be effective with higher local recurrence after breast-conserving therapy, the distant recurrence, breast cancer mortality, and death by any cause in patients with NAT were not statistically different from those with adjuvant therapy.7 The advantages of NAT for breast cancer include decreasing the tumor size, improving outcomes of radical or more conservative surgical interventions and early evaluation of clinical efficacy,8 and serving as an excellent research platform to test new predictive biomarkers by tumor and/or blood sampling prior to and during systemic therapy.6 However, tailored therapies based on clinical responses to standard NAT are still not well established because of the highly heterogeneous nature of breast cancer, which presents various subtypes at the molecular, histopathological, and clinical levels.9 Robust predictive biomarkers9,10 and reliable measures of clinical benefit from biomarker-derived personalized therapy remain limited.6 This work offers an overview of the literature related to biomarkers that may predict the response to NAT for breast cancer. These biomarkers provide clinical, biological, and imaging information.
Clinical characteristics
Clinical characteristics usually include clinical and demographic features, such as family history, menstrual status, patient’s age, mammographic breast density, and racial disparity. Family history that includes cases of early onset or bilateral breast cancers and multiple cases of breast or ovarian cancers can be a strong predictor of hereditary breast cancer.11 Menstrual status has become a pivotal consideration while selecting optimal endocrinal treatment strategies.6,12,13 Patient’s age is an important prognostic factor for patients positive for hormone receptors, but for patients positive for human epidermal growth factor receptor 2 (HER2+) or triple-negative breast cancer (TNBC), age is not an independent prognostic factor.14 Moreover, mammographic breast density might be associated with response to NAT and a low mammographic density could predict improved pathological complete remission (pCR).15 In addition, the response to NAT displays racial disparity, wherein HER2+ metastatic breast cancer presented worse clinical outcomes when comparing Caucasian with African-American patients.16 Whether these clinical and demographic features can robustly predict the response to NAT warrants further investigation. This article does not contain any individual participants that required informed consent. This article does not contain any studies with human participants or animals performed by any of the authors.
Pathological features
Pathological features of breast cancer may include the histological tumor type and grade; tumor-node-metastasis (TNM) stage; tumor estrogen receptor (ER), progesterone receptor (PR), and HER2 status; Ki-67 index; mitotic counts; and necrosis. According to the status of ER, PR, HER2, and Ki-67, breast cancer was divided into four different molecular subtypes, such as luminal A, luminal B, HER2+, and TNBC.6 Each subtype might have different treatment strategies, and the predictive factors may not be the same. For example, the best treatment regimen for breast cancer with luminal A subtype, which is clinically acceptable, is endocrine therapy alone without chemotherapy,17 whereas for luminal B subtype, the optimal combination of chemotherapy and endocrine therapy is necessary. Recently, the PAMELA study showed that breast cancer patients with the HER2+ subtype are more likely to benefit from the dual HER2 blockade treatment.18 TNBC itself represents a heterogeneous group with varying treatment sensitivities and prognoses. Various systemic therapies including those with cytotoxic and molecularly targeted agents improve the response rate of NAT for breast cancer.19
Classical morphological parameters, such as lobular histotype and presence of inflammation, predict the response to NAT, particularly in luminal B and HER2+ subgroups.20 The pattern of central necrosis and fibrosis has been found to affect the prognosis and prediction of breast cancer. In a pooled data analysis, the outcome in the presence of central necrosis and fibrosis was worse than those in their absence, regardless of tumor grade. In contrast, in the absence of central necrosis and fibrosis, patients with grade 3 breast cancer tumors had poorer outcome than those with grade 1–2 tumors.21 Mitosis counting and Ki-67 index, which are proliferation markers of tumor cells, have proven effective, practical, easily assessable, inexpensive, and highly reproducible prognostic factors and predictors. Higher mitotic counts were associated with the presence of lymph node metastasis.22 In an NAT cohort study, histological grade was not found to be significantly correlated with Ki-67, even though, in approximately two-thirds of the cases, Ki-67 decreased after NAT.23 Some of these pathological features have proven to be predictive, eg, ER and HER2. However, their predictive response to NAT warrants further clinical validation.
Pathological complete response (pCR), which was defined as the absence of tumor cells in the surgical specimen, both at the primary tumor site and at regional lymph nodes,24–26 has been used as an endpoint in numerous trials of neoadjuvant systemic therapy for breast cancer, and molecular subtypes have been independently associated with pCR rate.27 In the Collaborative Trials in Neoadjuvant Breast Cancer (CTneoBC) pooled analysis,25 which assessed the relationship between pCR and long-term outcome, the absence of residual invasive cancer in the breast and axillary nodes with the presence or absence of in situ cancer provided a better association with improved outcomes than eradication of invasive tumor from the breast alone. Complete remission both in breast and lymph nodes, especially in patients with TNBC and in those with HER2-positive breast cancer, has been proposed as a surrogate endpoint for the prediction of long-term clinical benefit, such as disease-free survival, event-free survival (EFS), and overall survival (OS).25,28,29 Some pathological features, such as smaller tumor size, absence of axillary lymph nodes,4,30 and histological grade 3,31–33 are more likely to achieve pCR.
Circulating biomarkers
Circulating markers include soluble cells, molecules, and exosomal nucleic acids.34 They might be useful for diagnosis, prognosis, and real-time therapy monitoring with lower costs and higher compliance than tumor biopsy, due to their minimal invasiveness.34 This section addresses circulating tumor cells (CTCs),35 circulating endothelial cells (CECs),36 and the circulating tumor DNA (ctDNA), which are rare populations in the peripheral circulation and potential biomarkers. Although myeloid-derived suppressor cells (MDSCs) and various lymphocyte subpopulations are also present in the peripheral blood, they are discussed in the “Immunological biomarkers” section.
CTCs
The presence of disseminated tumor cells in the bone marrow, as well as detection and persistence of CTC, enables the identification of patients with especially poor prognosis. In patients with metastatic breast cancer, the presence and enumeration of CTC in the peripheral blood are associated with reduced survival.37–39 Elevated CTC levels in the early course of treatment were found to be the early independent marker, predictive of poor survival,40 while molecular profiling of CTC offers superior prognostic information for assessing the risk of recurrence and superior predictive judgment of therapeutic regimens.41 In adjuvant42–44 or neoadjuvant45,46 settings, CTCs could also be prognostic factors, which suggests that they could potentially be used as a monitoring tool during follow-up and a selection criterion of patients for secondary treatment intervention. For inflammatory breast cancer treated with NAT, CTC detection at the baseline was associated with 3-year shorter disease-free survival and OS. This suggests that CTC could be used for prediction and stratification.47 Recently, a meta-analysis with individual patient data from 21 studies showed that CTC was detectable in 25.2% of patients before NAT and the number of CTCs could negatively affect the OS, distant disease-free survival, and locoregional relapse-free interval, which suggest that the CTC count is an independent and quantitative prognostic factor for the treatment of breast cancer with NAT.48 The challenges associated with CTC detection and characterization include its scarcity, difficult enrichment, and limited blood volume for analysis.49 A reliable method for detecting CTCs warrants further improvement,50 and their value for predicting response to NAT needs to be validated in clinical trials.
ctDNA
Compared with healthy people, cancer patients usually have higher levels of circulating cell-free DNA (cfDNA).51 The ctDNA harbors DNA mutations, epigenetic alterations, and other forms of tumor-specific abnormalities.52 It is released into the bloodstream both from tumor tissue and from lysed CTCs following apoptosis and/or necrosis of tumor cells.51,53 ctDNA can be detected in various solid malignancies, and its levels are associated with the disease stage.
Various genetic and epigenetic events including DNA strand integrity, gene amplifications, gene mutations, DNA methylation, and microsatellite abnormalities might occur during the initiation and progression of breast cancer. Studies on circulating DNA in plasma and serum of patients with breast cancer are well summarized.54 These studies have suggested that analyses of circulating DNA might provide prognostic and predictive information for the diagnosis and treatment of breast cancer.54 With the RNA sequencing, high levels of HER2 and low levels of ESR1 (ER 1) were shown to be associated with higher pCR for breast cancer.55 However, the role of cfDNA or ctDNA in the prediction of the response to NAT has not been demonstrated. Analysis of cfDNA is performed using technologies such as quantitative polymerase chain reaction (qPCR); beads, emulsion, amplification, and magnetics (BEAMing); pyrophosphorolysis-activated polymerization (PAP); combined bisulfite restriction analysis (COBRA); and microarray and next-generation sequencing among others. Each of these techniques presents various advantages and limitations.54
CECs
Endothelial cells can enter circulation as CECs when sloughing off vessel walls and as endothelial progenitor cells (EPCs) when mobilized from the bone marrow.36 Both CECs and their populations, which can be enumerated by flow cytomery,56–58 could potentially serve as biomarkers for the prognosis and prediction of various solid tumors.36 CECs and EPCs appear to change dynamically during the course of chemotherapy of patients with breast cancer. A decrease in CECs and an increase in EPCs could serve as surrogate markers of angiogenesis in antiangiogenesis treatments combined with chemotherapy.59 For patients with metastatic breast cancer, orally administered S-1 could suppress CECs, as measured using the CellSearch system.60 In adjuvant settings, CEC numbers could predict higher Nottingham prognostic index scores and correlated positively with tumor invasiveness and size, which might reflect total tumor vascular volume.61 In neoadjuvant settings, 35 patients with operable breast cancer received NAT with a regimen based on anthracycline and/or taxane. The number of baseline CEC, in particular CD34+ CEC, and the percentage of CD34+ cells were associated with the rate of pCR of NAT, which suggests that CEC could potentially be used as a predictive biomarker for NAT.62 However, the extent and the significance of the contribution of CECs and EPCs for breast cancer growth are still not well defined.63
CTCs, CECs and their subsets, and ctDNA are “magic tools” of liquid biopsy, promising new biomarkers in oncology, with potential clinical applications for the monitoring and comprehensive molecular profiling of breast cancer.64 Great challenges remain in the interpretation of the data and their optimal utilization in improving patient treatment and outcomes.65
Biochemical markers
A variety of biochemical biomarkers, including proteins, enzymes, DNA, and RNA, could be used as predictive biomarkers for detecting breast cancer and monitoring patient’s treatments. Glycosylated proteins such as carcinoembryonic antigen (CEA), cancer antigen 15-3 (CA15-3), and cancer antigen 125 (CA125) are biochemical markers, whereas ER, PR, HER2, and Ki-67 are biomolecular markers.
CA15-3 and CEA, the biomarkers most frequently used in clinical practice, are serum-based glycosylated tumor markers that have been approved by the US Food and Drug Administration (FDA) for monitoring breast cancer.66,67 Several studies have shown that elevated levels of serum CA15-3 and CEA may be used for detection, prognostic estimation, and treatment response prediction in patients with breast cancer.68–70 Moazzezy et al69 reported that the serum levels of CA15-3 and CEA were independent of the stage of the breast cancer. A study by Uehara et al67 found significant differences in the expression level of CA15-3 across different stages of breast cancer.67 Samy et al71 showed that preoperative serum levels of CEA in breast cancer patients were significantly higher than those in the control group. A meta-analysis that included 13 case–control studies showed that both CA15-3 and CEA showed potential as biomarkers for breast cancer monitoring and were strongly associated with the clinical stage.72 In addition, tumor marker CA125 and malignant tumor-specific growth factor were shown to be differentially expressed in breast cancer patients compared to the control subjects. However, in the breast cancer guideline from American Society of Clinical Oncology (ASCO),73 CEA, CA15-3, CA27.29, lactate dehydrogenase, and others were not recommended for the screening, diagnosis, staging, or routine surveillance of breast cancer patients after primary therapy, mainly because of their limited sensitivity and specificity, even though they could be minimally complementary to other clinical information.74
In addition, some emerging novel biomarkers such as noncoding RNA including microRNAs and long noncoding RNAs have been widely investigated for the diagnosis, prediction, and prognosis of breast cancer.75,76 Their detection, quantification, and molecular characterization have provided new avenues for studying the metastatic process and have provided new perspectives in terms of the early detection and prediction of breast cancer. A number of studies77–82 have investigated the predictive and/or prognostic value of circulating RNA in patients with breast cancer, but these results were inconclusive partially because of the limited patient number in the published studies. A number of issues regarding the normalization and standardization of sample collection, as well as measuring methods, warrant further improvement. In addition, the predictive response value of these noncoding RNA needs to be validated in a wide clinical setting.
Multigene/multiprotein profiling
Multigene profiling assays have been developed that might improve the prediction of outcomes compared to standard clinical and pathological markers. Biomarker assays such as Oncotype DX, MammaPrint, EndoPredict, PAM50, and Breast Cancer Index (BCI) could prove useful for specific subgroups of breast cancer.10 For example, if a patient has an ER/PR-positive HER2-negative breast cancer, 21-gene recurrence score (Oncotype DX; Genomic Health, Redwood City, CA, USA) can be used to guide decisions on adjuvant systemic chemotherapy.10,83 This multigene model showed 10-year recurrence rates among low-, intermediate-, and high-risk groups of 6.8, 14.3, and 30.5%, respectively.83 It is valuable to predict the probability of late recurrence and the magnitude of chemotherapy benefit, which could spare unnecessary adjuvant chemotherapy.84–87
MammaPrint, developed by the Netherlands Cancer Institute group, considers 70 genes related to early risk of metastasis, including tumor invasion-, metastasis-, interstitial invasion-, and angiogenesis-related genes.88,89 According to gene expression levels, the good- and poor-prognosis signatures showed significantly different risks of 10-year distant metastasis (85 and 51%) and OS (95 and 55%).89 Adjuvant chemotherapy could significantly reduce the rate of 10-year distant metastasis for breast cancer patients with poor-prognosis signature.90 Recently, the prospective MINDACT study91 confirmed the clinical utility of MammaPrint. Patients with early-stage breast cancer who are at a high clinical risk and low genomic risk for recurrence, based on the 70-gene signature, might not benefit from adjuvant chemotherapy, and 46% of breast cancer patients with a high clinical risk could be spared from chemotherapy.91 MammaPrint has been recommended to determine the prognosis and guide decision making for selecting patients who could benefit from adjuvant chemotherapy for invasive breast cancers that are lymph node negative or 1–3 lymph node positive.92
The Prosigna test (previously known as PAM50; NanoString Technologies, Seattle, WA, USA)93,94 measures the expression of 50 genes at the mRNA level, the EndoPredict test95 detects the expression levels of 11 genes (Sividon Diagnostics GmbH, Cologne, Germany), and the BCI (bioTheranostics, San Diego, CA, USA) test measures the expression of 11 genes.96 In combination with clinical and pathological factors, Prosigna, EndoPredict, and BCI have been recommended for predicting the outcome and aid decision making concerning adjuvant therapy in ER/PR-positive HER2-negative patients with negative or 1–3 positive lymph nodes.92
Although published guidelines give conflicting recommendations regarding breast cancer biomarkers,92 various multigene panels could improve risk discrimination relative to clinicopathological factors and the prediction of clinical outcome. Furthermore, they could identify patients who might safely avoid adjuvant chemotherapy, thereby saving costs, and could promote the progress of personalized clinical decision making. It has been recommended that TNM classification of breast cancer, described in the “Imaging biomarkers” section, is complemented with some multigene panels, such as Oncotype DX.33 However, the efficacy of multigene panels in predicting relapse rates in HR-positive breast cancer beyond 5 years remains to be investigated. Multigene panels were only validated in adjuvant settings; hence, they should be used with caution when extrapolating to a NAT context.
Immunological biomarkers
The immune system plays a pivotal role in maintaining tissue homeostasis by continuous immunosurveillance and the initiation of inflammatory reactions.97 Cancer, as it transforms from normal tissues, induces innate immune responses to eliminate incipient tumor cells via immunoediting.98,99 Tumor microenvironment within tumor regions, such as nontransformed elements, include immune cells or molecules, blood vessels, fibroblast, mesenchymal cells, adipocyte, and extracellular matrix, among others.100 Immune cells that contribute to tumor immunoediting include tumor-infiltrating lymphocytes (TILs), tumor-associated macrophages (TAMs), regulatory T cells (Tregs), type II natural killer T (NKT) cells, and MDSCs.101 Strategies are rapidly emerging that target various immune cells and tumor microenvironment to improve cancer treatment.102 Therefore, both tumor immunological status and microenvironment could potentially be used as biomarkers to predict response to anticancer therapy.
TILs, as immune cells that infiltrate tumor tissue, are present in various solid tumors including breast cancer.103 TILs, classified as either stromal or intratumoral, are usually detected in hematoxylin and eosin-stained histological slides.104 Many studies showed a positive association between higher TIL numbers and more favorable prognosis or better response to treatment.105 The presence of TILs at the time of diagnosis provides prognostic and potentially predictive values, particularly in cases of triple-negative and HER2 breast cancer patients.103–107 In adjuvant settings, TILs might serve as predictive biomarkers for the benefit from cytotoxic chemotherapy,108,109 while, in the neoadjuvant context, the presence of TILs has been found to be associated with increased rates of pCR.107 In a recent pooled analysis, the increased TIL concentration could predict a better response to NAT for all the molecular subtypes of breast cancer and a better survival for HER2+ and TNBC subgroups.110 TILs might also play an important role in immunomodulatory effects of chemotherapy. For example, chemotherapy might relieve immunosuppression and reset the attenuated functional immunity that was triggered by chemotherapy-induced cell death, thus possibly contributing to the elimination of the remaining primary viable tumor cells and distant micrometastatic disease.103 Nevertheless, a standardized methodology to measure TILs is needed to improve the consistency and reproducibility of future studies.104 In summary, understanding and exploiting the immunobiology of breast cancer, especially the importance of TILs in a breast cancer immune microenvironment, remain a great challenge.103
TAMs are prominent components of tumor microenvironment in breast cancer.111,112 Macrophages are usually polarized into classically activated (M1) and alternatively activated (M2) macrophages.113 TAMs demonstrate high plasticity in response to various signals and participate in the immune responses that control the tumor microenvironment.111,112 They are versatile and have been involved in the regulation of tumor growth, angiogenesis, invasion, metastasis, immunosuppression, and chemotherapeutic resistance.100,111,112 TAMs could also be used as important prognostic or predictive factors.100 High density of TAMs as a predictor of patient outcome after chemotherapy for various tumors has been well summarized.112 A recent meta-analysis of 16 studies with 4,541 patients showed that a high density of TAMs was related to worse OS and disease-free survival.114 High infiltration of TAMs correlated significantly with many clinicopathological features, such as age, tumor size, histological grade, ER/PR status, and vascular invasion.114 Because patient-to-patient variability in cancer progression complicates clinical treatment decisions, a multivariate predictive in vitro model, using monocyte-derived macrophages and related biomolecules, has been established, which allows the scoring of patients for invasion risk. This might prove useful to inform oncologists and patients of invasive/metastatic risk.115 Nevertheless, this predictive model should be validated for application to current breast cancer patients with long-term clinical outcomes in the context of a more robust dataset. Hopefully, TAMs will provide tools to tailor the use of local and systemic therapies in a personalized medicine approach, including chemotherapy, hormonal therapy, antiangiogenesis, and immunotherapy. TAM-focused therapeutic strategies might potentially complement and synergize with other cancer therapies.112 However, the biomarker potential of TAMs and the approach to target macrophages in cancer warrant further investigation.
MDSCs affect the local tumor microenvironment by suppressing host immune responses116,117 and by playing an important role in tumor formation and progression.101 Available treatment strategies that target MDSCs include inhibition of MDSC development, expansion, and function, as well as differentiation, depletion, or destruction of MDSCs.101 Several studies have indicated that the number of MDSCs in the peripheral blood of some cancer patients correlated with the clinical stage and metastatic tumor burden.101 In a case cohort study, myeloid cells were found to be significantly expanded in the tumor microenvironment of breast cancer patients.118 Circulating MDSCs have been found to be significantly higher in breast cancer patients compared to healthy volunteers, with the number of MDSCs correlating significantly with the clinical stage.119 In the setting of neoadjuvant chemotherapy for breast cancer, peripheral blood levels of granulocytic MDSCs increased significantly in all molecular subtypes during doxorubicin and cyclophosphamide therapy. Granulocytic MDSC levels were quantitatively lower in patients with pCR than in patients with no pCR.120 At present, the major problem in using MDSCs as a prognostic or predictive biomarker and their utilization in cancer research lie in the difficulty of reliably defining MDSCs themselves.
Lymphocyte subpopulations participate in distinct components of the immune response in various conditions, including solid tumors and leukemia.121 The total number of lymphocytes and the composition of lymphocyte subpopulations can be measured by flow cytometry.122,123 These lymphocyte subpopulations could be potentially used as predictive and prognostic biomarkers for many tumors, including breast cancer. The absolute number of regulatory T cells, characterized by the expression of CD4+CD25+CD127–, has been found to be higher in breast cancer patients compared to healthy controls, with numbers higher in patients with stage III or IV breast cancer than in those with stage I or II.124 The density of regulatory T cells that infiltrate intratumor before chemotherapy has been described as the strongest predictor for survival.125 For breast cancer, different lymphocyte subpopulations might respond differently to radiotherapy with or without chemotherapy. For example, the numbers of CD4(+) T cells and regulatory T cells increased during the treatment course of radiotherapy, while those of NK cells and B cells decreased. All numbers returned to normal within 6 months after radiation treatment.126 Elevated numbers of circulating CD8+CD28– suppressor T cells have been associated with shorter progression-free survival and OS for metastatic breast cancer.127,128 Depressed circulating anti-HER2 CD4-positive (CD4+) T-helper type 1 (Th1) response correlated with higher risk of recurrence in patients with completely treated HER2-positive invasive breast cancer129 and has been related to pathological response following NAT in HER2-positive breast cancer,130 which suggested that benefit from trastuzumab therapy may be restricted to immune-enriched tumors. Though several studies suggest that lymphocyte subpopulations in tumor or peripheral blood could potentially be used as biomarkers for breast cancer prediction and prognosis, it should be validated in prospective studies recruiting higher numbers of patients. Additionally, standardization of the measurement of each lymphocytic subpopulation should be improved.
Imaging biomarkers
Various breast imaging examinations are assessed prior to NAT to evaluate the extent of disease and axillary lymphadenopathy, as well as to screen the contralateral breast. Typically, patients undergo physical examination and breast imaging, such as mammography and/or ultrasonography (US), that have been proposed to evaluate response to NAT,131 but they were only moderately useful for predicting residual pathological tumor size after NAT.132 Physical examination, which has an accuracy of 57%, might overestimate the amount of residual disease due to the presence of firm fibroglandular tissue and post-therapy fibrosis.133 The diagnostic accuracy of mammography and US for determining the pCR rate of NAT was measured to be 74 and 79%, respectively.134 Breast ultrasound has been more accurate than mammography in predicting residual tumor size.135 When both imaging modalities demonstrated no residual disease, the likelihood of pCR was found to be 80%.135 The advantages and disadvantages of mammography and US have been well reviewed.133,135–137 In this section, the potential of molecular imaging such as positron emission tomography (PET)–computed tomography (CT), magnetic resonance (MR), and radiomics as biomarkers for breast cancer is discussed.
PET–CT imaging biomarker
Fluorine 18 fluorodeoxyglucose (F18-FDG) is the most commonly used molecular imaging tracer for imaging tumor metabolism by PET–CT. Imaging with PET–CT can be used, optionally, to determine the systemic stage of breast for cancer patients with stage III or IV of the disease or with disease failure after antitumor therapy.6
Several studies have been performed to evaluate the potential of F18-FDG PET–CT imaging to predict breast cancer pCR after NAT. In a cohort study with locally advanced breast cancer that received NAT, the standardized uptake value (SUV) in responding tumors F18-FDG decreased significantly just after the first course of chemotherapy. The SUV of all responders decreased <55% of their baseline, with a sensitivity of 100% and a specificity of 85%.138 Histopathological response could be predicted with an accuracy of 88–91% after the first or second cycle of NAT.138 Early changes in SUV after a course of NAT was a good independent biomarker to predict pathological response.139 Similarly, for patients with metastatic breast cancer, longitudinal PET might allow the prediction of response to treatment after the first cycle of chemotherapy.140 Three meta-analyses studying F18-FDG PET–CT for the evaluation of response to NAT showed a pooled sensitivity and specificity of 80–85 and 66–79%, respectively.133 Based on the available literature, F18-FDG PET–CT imaging could potentially be used as an early selection marker of chemosensitive patients. It could help identify ineffective chemotherapy and direct patients to either alternative therapy or surgery.133 The use of new tracers for PET–CT, such as fluorothymidine,141 radiolabeled essential amino acid,11,142 C-choline,143 and fluciclovine,144 has been investigated in a number of case-cohort studies to predict the response of breast cancer patients to NAT. Prospective, response-guided clinical trials are needed to validate these possible advantages.
MR imaging biomarker
MR, a powerful, versatile, and precise imaging technique, is the most sensitive modality for breast cancer detection145,146 and the most accurate imaging technique for assessing tumor response to NAT.147–150
Dynamic contrast-enhanced (DCE) MR, the backbone of the breast MR imaging protocol, has an excellent sensitivity and a good specificity for breast cancer detection.145,151 Pharmacokinetic analysis of DCE perfusion MR imaging has demonstrated that the volume transfer constant (Ktrans), which describes the trans-endothelial transport of imaging contrast agent from blood vessels into the extravascular–extracellular space by diffusion,131 is a promising parameter for the early identification of treatment response.152,153 Ktrans changes during the course of NAT and may predict pathological response and long-term survival.154,155 Due to quantification and reproducibility, it has been used widely in the clinical practice for breast cancer and standardized techniques and methodological analyses of DCE MR imaging quantification have been recommended.131,156
Diffusion-weighted (DW) MR imaging can be used to complement DCE MR imaging to evaluate tumor response to NAT. Compared with normal tissue, most invasive breast cancers have lower apparent diffusion coefficient (ADC) values.157 Anticancer agents usually result in cell membrane damage and reduce the number of viable malignant cells. Therefore, ADC values tend to increase more significantly in pathological responders than in nonresponders after chemotherapy.158,159 Similar to Ktrans, ADC value may change after only one cycle of chemotherapy and before the tumor size is affected. A meta-analysis,160 with 34 studies covering 1,932 breast cancer patients, reported the sensitivity and specificity of DW MR to predict the pCR of 0.93 and 0.82, respectively. In a recent meta-analysis of 57 published studies, >80% of studies used the DCE MR technique and most studies used a 1.5 T magnet strength to evaluate pCR of the patients who received NAT.150 The pCR rate ranged from 4.1 to 54.9% with a median of 16.8%. Sensitivity and specificity (correct detection of residual tumor) of DCE MR were reported to be 0.64 and 0.92, respectively, while those of DW MR were 0.93 and 0.85, respectively. The accuracy rate, calculated from Youden’s index, of DCE MR and DW MR was similar.150 DCE MR had greater accuracy than US and mammography and PET–CT or DW MR had high sensitivity, and DCE MR had high specificity.150
Proton MR spectroscopy allows the evaluation of total choline (tCho) peak as a biomarker of breast cancer.161 The resonance peak of tCho might decrease or disappear completely in locally advanced breast cancer patients undergoing chemotherapy.162 In breast cancer patients undergoing NAT, tCho decreased163 more significantly within 24 hours of initial treatment in responders than in nonresponders. Levels of tCho164 reduced even more after 1–2 cycles of chemotherapy. In addition, diffusion tensor imaging (DTI), an extension of standard DW with diffusion encoding, can measure the full diffusion tensor and characterize the motion of water.165 In a case cohort, DTI was used to monitor the changes in volume and diffusion tensor parameters were proven to predict response to NAT as accurately as DCE.165,166
Most of the MR imaging studies149,150,165–167 were performed on relatively small cohorts, which might limit the statistical power and warrant further validation and refinement in large prospective research projects. In addition, color ultrasonic studies might provide more valuable information when combined with MR functional imaging such as DW, DCE, spectroscopy MR, and some other multimodal imaging, for example, PET–CT with various tracers. Both the molecular subtype of breast cancer and the antitumor regimen might influence the accuracy of breast MR detection of residual lesions.147 In addition, both DW MR and DTI MR are research tools with major limitations, such as relatively long acquisition times; frequent low quality spectra; and difficulty in standardization, quantification of tCho tissue concentration, and fat suppression.133,163–165
Radiomics biomarker
Radiomics is a bridge between medical imaging and personalized medicine via high-throughput data mining. It enables image features to be quantified and used for cancer diagnosis and treatment, which could improve diagnostics, prognostics, and predictive accuracies.168 With quantitative computer-aided radiomic techniques, the tumor and its surrounding parenchyma could present thousands of imaging features including size, shape, margin morphology, enhancement texture, and kinetic assessment. These features could help identify the molecular subtypes of breast cancer.169 Radiomics features provide a novel imaging biomarker for estimating intratumoral heterogeneity.170 Intratumoral texture features have the potential to serve as a valuable imaging biomarker to enhance the prediction of breast cancer prognosis.171 Metabolic radiomic patterns in breast cancer have been associated with the propensity of pCR after NAT and risk of recurrence,172 which suggests that they could serve as biomarkers with predictive and prognostic abilities for the personalized management of breast cancer. For patients with invasive breast cancer, the radiomics signature on MR has proved to be an independent biomarker for the estimation of disease-free survival.173
Challenges and future directions
Response evaluation criteria of NAT and clinical validation
For the majority of solid cancers, changes in tumor burden are usually evaluated using validated and consistent criteria, such as response evaluation criteria in solid tumors (RECIST),174 immune-related response RECIST,175 and PET response criteria in solid tumors (PERCIST).176 The objective of NAT might be slightly different from the adjuvant or palliative contexts. Therefore, these response evaluation criteria should be used with caution when extrapolating to neoadjuvant settings. Although the use of various proteins and molecules such as miRNAs and exosomes as novel biomarkers has opened new opportunities for cancer diagnosis and monitoring, we are now facing several challenges including documentation of test reproducibility in multiple clinical trials, standardization of data acquisition, data analysis, reporting of results, and demonstration of strong correlations between test and clinical outcomes. In addition, these biomarkers need to be widely validated in well-designed, biomarker-guided, prospective trials before their final clinical application. In the era of precision medicine and big data, a closer integration of breast biology, immunology, imaging, and related biomedical fields is necessary. In addition, the creation of large integrated and shareable databases of clinical, molecular, and imaging biomarkers could promote advancement in the field to continue guiding breast cancer care and research.177
Combined multimodality biomarker approaches
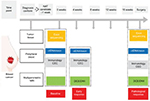
Combining quantitative imaging features with multigene assays could provide a promising mean for image-based phenotyping to assess the risk of breast cancer recurrence.178 At present, >1,300 trials have been registered to investigate potential biomarkers for breast cancer. We have also initiated a clinical trial (clinicaltrials.gov identifier: NCT03242551) to investigate the potential value of combining biomarkers from breast tumor, peripheral blood, and imaging (Figure 1). Merging these clinical characteristics, imaging phenotypes, and genomic data may lead to improved clinical outcome predictors. Reliable, quantitative prognostic or predictive models with multiple parameters of breast cancer could prove useful for precision medicine and for deciding on a patient treatment strategy. Radiogenomics, which integrates imaging characteristics with gene expression patterns, gene mutations, and other genome-related characteristics, will facilitate a deeper understanding of tumor biology and will enable the capturing of intrinsic tumor heterogeneity. 9
Predictive biomarkers for the molecular target therapy or immunotherapy
Both molecular target agents and immune checkpoint inhibitors are emerging as an important solution for recurrent or metastatic breast cancer, and some of these agents are increasingly used in the adjuvant or neoadjuvant setting. Most of the novel agents target one or more molecular pathways that affect the tumor growth and/or its microenvironments. For the early-stage breast cancer, some patients might be resistant to the aromatase inhibitor such as anastrozole and the cyclin-dependent kinase (CDK) 4/6 inhibitors, eg, palbociclib, which could be an active antiproliferative agent.180 The combination of an mTOR pathway inhibitor such as everolimus with chemotherapy could be used in the NAT of TNBC.181 The combination of immunotherapy agent and HER2 inhibitors is promising in advanced breast cancer patients, but the efficacy of immune agents in NAT or in an adjuvant setting has not been shown.182 Though these new treatment agents are increasingly available and widely used, the response and survival benefit as well as the reliable biomarkers that might stratify response to NAT need to be further validated.
Conclusion
Despite massive efforts toward the development of novel biomarkers for breast cancer, only three, ie, ER, PR, and HER2, biomarkers are recommended as mandatory in international guidelines.6,10,12,92 Reliable biomarkers currently used in clinical practice include ER and PR for predicting the benefit from endocrine therapy and HER2 for predicting the benefit from anti-HER2 therapy. A few clinical features, such as age and menstrual status, are useful to make clinical decisions and could predict the risk of failure after anticancer therapy and guide the selection of treatment strategies. Some multigene assays, for example, Oncotype DX, MammaPrint, and Prosigna, could be recommended for specific subgroups of early-stage breast cancer patients. A few imaging biomarkers, such as SUV in PET–CT and Ktrans and ADC in MR, as well as their changes during the course of NAT, could be used to identify patients not benefiting from the treatment and to direct them to continue or change the delivery of NAT. Although biochemical biomarkers from peripheral blood and/or tumor tissue appear promising, there is still a lack of consensus in practice guidelines to guide the NAT of breast cancer. In addition, the amount of circulating cancer biomarkers and their profiles are growing almost exponentially, which might prove useful in discriminating among different molecular cancer subtypes. A reliable technical and clinical quality of biomarker assays needs to be established. Standardized reporting, interpretation of results, and assessment of interlaboratory variation should be carried out.183 The clinical utility of biomarker-derived NAT for breast cancer still requires further evidence in prospective multicenter clinical trials, particularly in the context of big sharable databases.
Acknowledgments
We would like to acknowledge Editage Inc. for careful English proofreading. This work was funded by the Sanming Project of Medicine in Shenzhen (nos SZSM201612023 and SYLY201724) and Shenzhen Clinical research funding (2018A01008).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. | ||
Peart O. Breast intervention and breast cancer treatment options. Radiol Technol. 2015;86(5quiz 559-562.):535M-–5558. | ||
Cianfrocca M, Goldstein LJ. Prognostic and Predictive Factors in Early-Stage Breast Cancer. Oncologist. 2004;9(6):606–616. | ||
Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. | ||
Kaufmann M, von Minckwitz G, Mamounas EP, et al. Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Annals of Surgical Oncology. 2012;19(5):1508–1516. | ||
Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018;19(1):27–39. | ||
Rubovszky G, Horváth Z. Recent Advances in the Neoadjuvant Treatment of Breast Cancer. J Breast Cancer. 2017;20(2):119–131. | ||
Kaufmann M, Pusztai L. Use of standard markers and incorporation of molecular markers into breast cancer therapy: Consensus recommendations from an International Expert Panel. Cancer. 2011;117(8):1575–1582. | ||
Harris LN, Ismaila N, Mcshane LM, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34(10):1134–1150. | ||
Brandt A, Lorenzo Bermejo J, Sundquist J, Hemminki K. Breast cancer risk in women who fulfill high-risk criteria: at what age should surveillance start? Breast Cancer Res Treat. 2010;121(1):133–141. | ||
Coates AS, Winer EP, Goldhirsch A, et al. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26(8):1533–1546. | ||
Curigliano G, Burstein HJ, P Winer E. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700–1712. | ||
Loibl S, Jackisch C, Lederer B, et al. Outcome after neoadjuvant chemotherapy in young breast cancer patients: a pooled analysis of individual patient data from eight prospectively randomized controlled trials. Breast Cancer Res Treat. 2015;152(2):377–387. | ||
Elsamany S, Alzahrani A, Abozeed WN, et al. Mammographic breast density: Predictive value for pathological response to neoadjuvant chemotherapy in breast cancer patients. Breast. 2015;24(5):576–581. | ||
Rugo HS, Brufsky AM, Ulcickas Yood M, et al. Racial disparities in treatment patterns and clinical outcomes in patients with HER2-positive metastatic breast cancer. Breast Cancer Res Treat. 2013;141(3):461–470. | ||
Gao JJ, Swain SM. Luminal A Breast Cancer and Molecular Assays: A Review. Oncologist. 2018;23(5):556–565. | ||
Llombart-Cussac A, Cortés J, Paré L, et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18(4):545–554. | ||
Loibl S, O’Shaughnessy J, Untch M, et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol. 2018;19(4):497–509. | ||
Balmativola D, Marchiò C, Maule M, et al. Pathological non-response to chemotherapy in a neoadjuvant setting of breast cancer: an inter-institutional study. Breast Cancer Res Treat. 2014;148(3):511–523. | ||
Maiorano E, Regan MM, Viale G, et al. Prognostic and predictive impact of central necrosis and fibrosis in early breast cancer: Results from two International Breast Cancer Study Group randomized trials of chemoendocrine adjuvant therapy. Breast Cancer Res Treat. 2010;121(1):211–218. | ||
Buhmeida A, Al-Maghrabi J, Merdad A, et al. Prognostic value of mitotic counts in breast cancer of Saudi Arabian patients. Anticancer Res. 2011;31(1):97–103. | ||
Petric M, Martinez S, Acevedo F, et al. Correlation between Ki67 and Histological Grade in Breast Cancer Patients Treated with Preoperative Chemotherapy. Asian Pacific Journal of Cancer Prevention. 2015;15(23):10277–10280. | ||
Macchia G, Gambacorta MA, Masciocchi C, et al. Time to surgery and pathologic complete response after neoadjuvant chemoradiation in rectal cancer: A population study on 2094 patients. Clin Transl Radiat Oncol. 2017;4:8–14. | ||
Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. | ||
Cortazar P, Geyer CE. Pathological Complete Response in Neoadjuvant Treatment of Breast Cancer. Ann Surg Oncol. 2015;22(5):1441–1446. | ||
Houssami N, Macaskill P, von Minckwitz G, Marinovich ML, Mamounas E. Meta-analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur J Cancer. 2012;48(18):3342–3354. | ||
von Minckwitz G, Untch M, Blohmer JU, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30(15):1796–1804. | ||
Mazouni C, Peintinger F, Wan-Kau S, et al. Residual Ductal Carcinoma In Situ in Patients With Complete Eradication of Invasive Breast Cancer After Neoadjuvant Chemotherapy Does Not Adversely Affect Patient Outcome. Journal of Clinical Oncology. 2007;25(19):2650–2655. | ||
Fung F, Cornacchi SD, Vanniyasingam T, et al. Predictors of 5-year local, regional, and distant recurrent events in a population-based cohort of breast cancer patients. Am J Surg. 2017;213(2):418–425. | ||
Rakha EA, El-Sayed ME, Menon S, Green AR, Lee AHS, Ellis IO. Histologic grading is an independent prognostic factor in invasive lobular carcinoma of the breast. Breast Cancer Res Treat. 2008;111(1):121–127. | ||
Alvarado-Cabrero I, Alderete-Vázquez G, Quintal-Ramírez M, Patiño M, Ruíz E. Incidence of pathologic complete response in women treated with preoperative chemotherapy for locally advanced breast cancer: correlation of histology, hormone receptor status, Her2/Neu, and gross pathologic findings. Ann Diagn Pathol. 2009;13(3):151–157. | ||
Giuliano AE, Connolly JL, Edge SB, et al. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA: A Cancer Journal for Clinicians. 2017;67(4):290–303. | ||
Ravelli A, Reuben JM, Lanza F, et al. Breast cancer circulating biomarkers: advantages, drawbacks, and new insights. Tumor Biology. 2015;36(9):6653–6665. | ||
Bielčiková Z, Jakabová A, Pinkas M, Zemanová M, Kološtová K, Bobek V. Circulating tumor cells: what we know, what do we want to know about them and are they ready to be used in clinics? Am J Transl Res. 2017;9(6):2807–2823. | ||
Zhou F, Zhou Y, Dong J, Tan W. Circulating endothelial cells and their subsets: novel biomarkers for cancer. Biomark Med. 2017;11(8):665–676. | ||
Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med Overseas Ed. 2004;351(8):781–791. | ||
Bidard FC, Vincent-Salomon A, Sigal-Zafrani B, et al. Prognosis of women with stage IV breast cancer depends on detection of circulating tumor cells rather than disseminated tumor cells. Ann Oncol. 2008;19(3):496–500. | ||
Budd GT, Cristofanilli M, Ellis MJ, et al. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12(21):6403–6409. | ||
Pierga JY, Hajage D, Bachelot T, et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann Oncol. 2012;23(3):618–624. | ||
Tewes M, Aktas B, Welt A, et al. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat. 2009;115(3):581–590. | ||
Falck A-K, Bendahl P-O, Ingvar C, et al. Analysis of and prognostic information from disseminated tumour cells in bone marrow in primary breast cancer: a prospective observational study. BMC Cancer. 2012;12(1):403. | ||
Domschke C, Diel IJ, Englert S, et al. Prognostic Value of Disseminated Tumor Cells in the Bone Marrow of Patients with Operable Primary Breast Cancer: A Long-term Follow-up Study. Ann Surg Oncol. 2013;20(6):1865–1871. | ||
Hartkopf AD, Wallwiener M, Fehm TN, et al. Disseminated tumor cells from the bone marrow of patients with nonmetastatic primary breast cancer are predictive of locoregional relapse. Ann Oncol. 2015;26(6):1155–1160. | ||
Hall C, Krishnamurthy S, Lodhi A, et al. Disseminated tumor cells predict survival after neoadjuvant therapy in primary breast cancer. Cancer. 2012;118(2):342–348. | ||
Mathiesen RR, Borgen E, Renolen A, et al. Persistence of disseminated tumor cells after neoadjuvant treatment for locally advanced breast cancer predicts poor survival. Breast Cancer Research. 2012;14(4): R117. | ||
Pierga JY, Bidard FC, Autret A. Circulating tumour cells and pathological complete response: independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Anna Oncol. 2017;28(1):103–109. | ||
Bidard FC, Michiels S, Riethdorf S, et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J Natl Cancer Inst. 2018;110(6):560–567. | ||
Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25(12):2304–2313. | ||
Jueckstock J, Rack B, Friedl TWP, et al. Detection of circulating tumor cells using manually performed immunocytochemistry (MICC) does not correlate with outcome in patients with early breast cancer – Results of the German SUCCESS-A- trial. BMC Cancer. 2016;16(1):401. | ||
Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: Practical aspects and biological significance. Mutation Research/Reviews in Mutation Research. 2007;635(2-3):105–117. | ||
Cai C, Guo Z, Cao Y, Zhang W, Chen Y. A dual biomarker detection platform for quantitating circulating tumor DNA (ctDNA). Nanotheranostics. 2018;2(1):12–20. | ||
Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. | ||
Schwarzenbach H, Pantel K. Circulating DNA as biomarker in breast cancer. Breast Cancer Research. 2015;17(1):136. | ||
Fumagalli D, Venet D, Ignatiadis M, et al. RNA Sequencing to Predict Response to Neoadjuvant Anti-HER2 Therapy: A Secondary Analysis of the NeoALTTO Randomized Clinical Trial. JAMA Oncol. 2017;3(2):227–234. | ||
Jacques N, Vimond N, Conforti R, et al. Quantification of circulating mature endothelial cells using a whole blood four-color flow cytometric assay. J Immunol Methods. 2008;337(2):132–143. | ||
Woywodt A, Blann AD, Kirsch T, et al. Isolation and enumeration of circulating endothelial cells by immunomagnetic isolation: proposal of a definition and a consensus protocol. J Thromb Haemost. 2006;4(3):671–677. | ||
Zhou F, Zhou Y, Yang M, Wen J, Dong J, Tan W. Optimized multiparametric flow cytometric analysis of circulating endothelial cells and their subpopulations in peripheral blood of patients with solid tumors: a technical analysis. Cancer Management and Research. 2018;10:447–464. | ||
Kuo Y-H, Lin C-H, Shau W-Y, et al. Dynamics of circulating endothelial cells and endothelial progenitor cells in breast cancer patients receiving cytotoxic chemotherapy. BMC Cancer. 2012;12(1):620. | ||
Tsuji W, Ishiguro H, Tanaka S, Takeuchi M, Ueno T, Toi M. Orally administered S-1 suppresses circulating endothelial cell counts in metastatic breast cancer patients. Int J Clin Oncol. 2014;19(3):452–459. | ||
Goon PK, Lip GY, Stonelake PS, Blann AD. Circulating endothelial cells and circulating progenitor cells in breast cancer: relationship to endothelial damage/dysfunction/apoptosis, clinicopathologic factors, and the Nottingham Prognostic Index. Neoplasia. 2009;11(8):771–779. | ||
Ali AM, Ueno T, Tanaka S, et al. Determining circulating endothelial cells using CellSearch system during preoperative systemic chemotherapy in breast cancer patients. Eur J Cancer. 2011;47(15):2265–2272. | ||
Botelho MC, Alves H. Endothelial Progenitor Cells in Breast Cancer. Int J Immunother Cancer Res. 2016;2:1–2. | ||
Gingras I, Salgado R, Ignatiadis M. Liquid biopsy: will it be the ‘magic tool’ for monitoring response of solid tumors to anticancer therapies? Curr Opin Oncol. 2015;27(6):560–567. | ||
Lowes L, Bratman S, Dittamore R, et al. Circulating Tumor Cells (CTC) and Cell-Free DNA (cfDNA) Workshop 2016: Scientific Opportunities and Logistics for Cancer Clinical Trial Incorporation. Int J Mol Sci. 2016;17(9):1505. | ||
O’Hanlon DM, Kerin MJ, Kent P, Maher D, Grimes H, Given HF. An evaluation of preoperative CA 15-3 measurement in primary breast carcinoma. Br J Cancer. 1995;71(6):1288–1291. | ||
Uehara M, Kinoshita T, Hojo T, Akashi-Tanaka S, Iwamoto E, Fukutomi T. Long-term prognostic study of carcinoembryonic antigen (CEA) and carbohydrate antigen 15-3 (CA 15-3) in breast cancer. Int J Clin Oncol. 2008;13(5):447–451. | ||
Park BW, Oh JW, Kim JH, et al. Preoperative CA 15-3 and CEA serum levels as predictor for breast cancer outcomes. Ann Oncol. 2008;19(4):675–681. | ||
Moazzezy N, Farahany TZ, Oloomi M, Bouzari S. Relationship between preoperative serum CA 15-3 and CEA levels and clinicopathological parameters in breast cancer. Asian Pac J Cancer Prev. 2014;15(4):1685–1688. | ||
Atoum M, Nimer N, Abdeldayem S, Nasr H. Relationships among serum CA15-3 tumor marker, TNM staging, and estrogen and progesterone receptor expression in benign and malignant breast lesions. Asian Pac J Cancer Prev. 2012;13(3):857–860. | ||
Samy N, Ragab HM, El Maksoud NA, Shaalan M. Prognostic significance of serum Her2/neu, BCL2, CA15-3 and CEA in breast cancer patients: A short follow-up. Cancer Biomarkers. 2010;6(2):63–72. | ||
Fu Y, Li H. Assessing Clinical Significance of Serum CA15-3 and Carcinoembryonic Antigen (CEA) Levels in Breast Cancer Patients: A Meta-Analysis. Med Sci Monit. 2016;22:3154–3162. | ||
Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312. | ||
Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312. | ||
Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. | ||
Amorim M, Salta S, Henrique R, Jerónimo C. Decoding the usefulness of non-coding RNAs as breast cancer markers. J Transl Med. 2016;14(1):265. | ||
Iorio MV, Casalini P, Tagliabue E, Ménard S, Croce CM. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer. 2008;44(18):2753–2759. | ||
Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic Significance of Metastasis-Related MicroRNAs in Early Breast Cancer Patients with a Long Follow-up. Clin Chem. 2014;60(1):197–205. | ||
D’Aiuto F, Callari M, Dugo M, et al. miR-30e* is an independent subtype-specific prognostic marker in breast cancer. Br J Cancer. 2015;113(2):290–298. | ||
Bailey ST, Westerling T, Brown M. Loss of Estrogen-Regulated microRNA Expression Increases HER2 Signaling and Is Prognostic of Poor Outcome in Luminal Breast Cancer. Cancer Res. 2015;75(2):436–445. | ||
Godinho MFE, Sieuwerts AM, Look MP, et al. Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br J Cancer. 2010;103(8):1284–1291. | ||
Chen Y-M, Liu Y, Wei H-Y, Lv K-Z, Fu P. Linc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cells. Tumor Biology. 2016;37(8):10861–10870. | ||
Paik S, Shak S, Tang G, et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. N Engl J Med Overseas Ed. 2004;351(27):2817–2826. | ||
Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. | ||
Sparano JA, Gray RJ, Makower DF, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N Engl J Med Overseas Ed. 2015;373(21):2005–2014. | ||
Albain KS, Barlow WE, Shak S, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. | ||
Gluz O, Nitz UA, Christgen M, et al. West German Study Group Phase III PlanB Trial: First Prospective Outcome Data for the 21-Gene Recurrence Score Assay and Concordance of Prognostic Markers by Central and Local Pathology Assessment. J Clin Oncol. 2016;34(20):2341–2349. | ||
van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. | ||
van de Vijver MJ, He YD, van ‘t Veer LJ, et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N Engl J Med Overseas Ed. 2002;347(25):1999–2009. | ||
Esteva FJ, Sahin AA, Cristofanilli M, et al. Prognostic Role of a Multigene Reverse Transcriptase-PCR Assay in Patients with Node-Negative Breast Cancer Not Receiving Adjuvant Systemic Therapy. Clinical Cancer Research. 2005;11(9):3315–3319. | ||
Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med Overseas Ed. 2016;375(8):717–729. | ||
Duffy MJ, Harbeck N, Nap M, et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer. 2017;75:284–298. | ||
Gnant M, Filipits M, Greil R, et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol. 2014;25(2):339–345. | ||
Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. | ||
Filipits M, Rudas M, Jakesz R, et al. A New Molecular Predictor of Distant Recurrence in ER-Positive, HER2-Negative Breast Cancer Adds Independent Information to Conventional Clinical Risk Factors. Clinical Cancer Research. 2011;17(18):6012–6020. | ||
Zhang Y, Schnabel CA, Schroeder BE, et al. Breast Cancer Index Identifies Early-Stage Estrogen Receptor-Positive Breast Cancer Patients at Risk for Early- and Late-Distant Recurrence. Clinical Cancer Research. 2013;19(15):4196–4205. | ||
Demaria S, Pikarsky E, Karin M, et al. Cancer and inflammation: promise for biologic therapy. J Immunother. 2010;33(4):335–351. | ||
Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells — mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14(3):155–167. | ||
Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases—elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. | ||
Choi J, Gyamfi J, Jang H, Koo JS. The role of tumor-associated macrophage in breast cancer biology. Histol Histopathol. 2017;11916. | ||
Anani W, Shurin MR. Targeting Myeloid-Derived Suppressor Cells in Cancer. Adv Exp Med Biol. 2017;1036:105–128. | ||
Wargo JA, Reuben A, Cooper ZA, Oh KS, Sullivan RJ. Immune Effects of Chemotherapy, Radiation, and Targeted Therapy and Opportunities for Combination With Immunotherapy. Semin Oncol. 2015;42(4):601–616. | ||
Savas P, Salgado R, Denkert C, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13(4):228–241. | ||
Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271. | ||
Simon RM, Paik S, Hayes DF. Use of Archived Specimens in Evaluation of Prognostic and Predictive Biomarkers. JNCI Journal of the National Cancer Institute. 2009;101(21):1446–1452. | ||
Seo AN, Lee HJ, Kim EJ, et al. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br J Cancer. 2013;109(10):2705–2713. | ||
Salgado R, Denkert C, Campbell C, et al. Tumor-Infiltrating Lymphocytes and Associations With Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated With Lapatinib and Trastuzumab. JAMA Oncol. 2015;1(4):448–454. | ||
Ali HR, Provenzano E, Dawson S-J, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12 439 patients. Annals of Oncology. 2014;25(8):1536–1543. | ||
Loi S, Sirtaine N, Piette F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31(7):860–867. | ||
Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19(1):40–50. | ||
Lewis CE, Pollard JW. Distinct Role of Macrophages in Different Tumor Microenvironments. Cancer Res. 2006;66(2):605–612. | ||
Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. | ||
Biswas SK, Allavena P, Mantovani A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin Immunopathol. 2013;35(5):585–600. | ||
Zhao X, Qu J, Sun Y, et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget. 2017;8(18):30576–30586. | ||
Park K-Y, Li G, Platt MO. Monocyte-derived macrophage assisted breast cancer cell invasion as a personalized, predictive metric to score metastatic risk. Sci Rep. 2015;5(1):13855. | ||
Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. | ||
Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32(1):19–25. | ||
Toor SM, Syed Khaja AS, El Salhat H, et al. Myeloid cells in circulation and tumor microenvironment of breast cancer patients. Cancer Immunology, Immunotherapy. 2017;66(6):753–764. | ||
Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin–cyclophosphamide chemotherapy. Cancer Immunology, Immunotherapy. 2009;58(1):49–59. | ||
Wesolowski R, Duggan MC, Stiff A, et al. Circulating myeloid-derived suppressor cells increase in patients undergoing neo-adjuvant chemotherapy for breast cancer. Cancer Immunology, Immunotherapy. 2017;66(11):1437–1447. | ||
Lebien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112(5):1570–1580. | ||
Brown JR, Wimberly H, Lannin DR, Nixon C, Rimm DL, Bossuyt V. Multiplexed Quantitative Analysis of CD3, CD8, and CD20 Predicts Response to Neoadjuvant Chemotherapy in Breast Cancer. Clinical Cancer Research. 2014;20(23):5995–6005. | ||
Mehr R, Sternberg-Simon M, Michaeli M, Pickman Y. Models and methods for analysis of lymphocyte repertoire generation, development, selection and evolution. Immunol Lett. 2012;148(1):11–22. | ||
Wang J, Yang J. Identification of CD4+CD25+CD127− regulatory T cells and CD14+HLA−DR−/low myeloid-derived suppressor cells and their roles in the prognosis of breast cancer. Biomed Rep. 2016;5(2):208–212. | ||
Demir L, Yigit S, Ellidokuz H, et al. Predictive and prognostic factors in locally advanced breast cancer: effect of intratumoral FOXP3+ Tregs. Clin Exp Metastasis. 2013;30(8):1047–1062. | ||
Sage EK, Schmid TE, Sedelmayr M, et al. Comparative analysis of the effects of radiotherapy versus radiotherapy after adjuvant chemotherapy on the composition of lymphocyte subpopulations in breast cancer patients. Radiother Oncol. 2016;118(1):176–180. | ||
Song Q, Ren J, Zhou X, et al. Circulating CD8 + CD28 − suppressor T cells tied to poorer prognosis among metastatic breast cancer patients receiving adoptive T-cell therapy: A cohort study. Cytotherapy. 2018;20(1):126–133. | ||
Song G, Wang X, Jia J, et al. Elevated level of peripheral CD8+CD28− T lymphocytes are an independent predictor of progression-free survival in patients with metastatic breast cancer during the course of chemotherapy. Cancer Immunology, Immunotherapy. 2013;62(6):1123–1130. | ||
Datta J, Fracol M, Mcmillan MT, et al. Association of Depressed Anti-HER2 T-Helper Type 1 Response With Recurrence in Patients With Completely Treated HER2-Positive Breast Cancer. JAMA Oncol. 2016;2(2):242–246. | ||
Datta J, Berk E, Xu S, et al. Anti-HER2 CD4+ T-helper type 1 response is a novel immune correlate to pathologic response following neoadjuvant therapy in HER2-positive breast cancer. Breast Cancer Research. 2015;17(1):71. | ||
O’Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14(3):169-186. | ||
Chagpar AB, Middleton LP, Sahin AA, et al. Accuracy of Physical Examination, Ultrasonography, and Mammography in Predicting Residual Pathologic Tumor Size in Patients Treated With Neoadjuvant Chemotherapy. Ann Surg. 2006;243(2):257–264. | ||
Fowler AM, Mankoff DA, Joe BN. Imaging Neoadjuvant Therapy Response in Breast Cancer. Radiology. 2017;285(2):358–375. | ||
Croshaw R, Shapiro-Wright H, Svensson E, Erb K, Julian T. Accuracy of Clinical Examination, Digital Mammogram, Ultrasound, and MRI in Determining Postneoadjuvant Pathologic Tumor Response in Operable Breast Cancer Patients. Ann Surg Oncol. 2011;18(11):3160–3163. | ||
Keune JD, Jeffe DB, Schootman M, Hoffman A, Gillanders WE, Aft RL. Accuracy of ultrasonography and mammography in predicting pathologic response after neoadjuvant chemotherapy for breast cancer. Am J Surg. 2010;199(4):477–484. | ||
Heine JJ, Malhotra P, Tissue M. breast cancer risk, serial image analysis, and digital mammography. Part 1. Tissue and related risk factors. Acad Radiol. 2002;9(3):298–316. | ||
Jafari SH, Saadatpour Z, Salmaninejad A, et al. Breast cancer diagnosis: Imaging techniques and biochemical markers. J Cell Physiol. 2018;233(7):5200–5213. | ||
Schelling M, Avril N, Nährig J, et al. Positron emission tomography using [18F]Fluorodeoxyglucose for monitoring primary chemotherapy in breast cancer. J Clin Oncol. 2000;18(8):1689–1695. | ||
Lee HW, Lee HM, Choi SE, et al. The Prognostic Impact of Early Change in 18F-FDG PET SUV After Neoadjuvant Chemotherapy in Patients with Locally Advanced Breast Cancer. J Nucl Med. 2016;57(8):1183–1188. | ||
Dose Schwarz J, Bader M, Jenicke L, Hemminger G, Jänicke F, Avril N. Early prediction of response to chemotherapy in metastatic breast cancer using sequential 18F-FDG PET. J Nucl Med. 2005;46(7):1144–1150. | ||
Kostakoglu L, Duan F, Idowu MO, et al. A Phase II Study of 3’-Deoxy-3’-18F-Fluorothymidine PET in the Assessment of Early Response of Breast Cancer to Neoadjuvant Chemotherapy: Results from ACRIN 6688. J Nucl Med. 2015;56(11):1681–1689. | ||
Lindholm P, Lapela M, Någren K, Lehikoinen P, Minn H, Jyrkkiö S. Preliminary study of carbon-11 methionine PET in the evaluation of early response to therapy in advanced breast cancer. Nucl Med Commun. 2009;30(1):30–36. | ||
Kenny LM, Contractor KB, Hinz R, et al. Reproducibility of [11C]choline-positron emission tomography and effect of trastuzumab. Clin Cancer Res. 2010;16(16):4236–4245. | ||
Ulaner GA, Goldman DA, Corben A, et al. Prospective Clinical Trial of 18F-Fluciclovine PET/CT for Determining the Response to Neoadjuvant Therapy in Invasive Ductal and Invasive Lobular Breast Cancers. J Nucl Med. 2017;58(7):1037–1042. | ||
Pinker K, Helbich TH, Morris EA. The potential of multiparametric MRI of the breast. Br J Radiol. 2017;90(1069):20160715. | ||
Mariscotti G, Houssami N, Durando M, et al. Accuracy of mammography, digital breast tomosynthesis, ultrasound and MR imaging in preoperative assessment of breast cancer. Anticancer Res. 2014;34(3):1219–1225. | ||
Lobbes MBI, Prevos R, Smidt M, et al. The role of magnetic resonance imaging in assessing residual disease and pathologic complete response in breast cancer patients receiving neoadjuvant chemotherapy: a systematic review. Insights Imaging. 2013;4(2):163–175. | ||
Yuan Y, Chen XS, Liu SY, Shen KW. Accuracy of MRI in prediction of pathologic complete remission in breast cancer after preoperative therapy: a meta-analysis. AJR Am J Roentgenol. 2010;195(1):260–268. | ||
Marinovich ML, Houssami N, Macaskill P, et al. Meta-analysis of magnetic resonance imaging in detecting residual breast cancer after neoadjuvant therapy. J Natl Cancer Inst. 2013;105(5):321–333. | ||
Yl G, Pan SM, Ren J, Yang ZX, Jiang GQ. Role of Magnetic Resonance Imaging in Detection of Pathologic Complete Remission in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy: A Meta-analysis. Clin Breast Cancer. 2017;17(4):245–255. | ||
Marino MA, Helbich T, Baltzer P, Pinker-Domenig K. Multiparametric MRI of the breast: A review. J Magn Reson Imaging. 2018;47(2):301-315. | ||
Marinovich ML, Sardanelli F, Ciatto S, et al. Early prediction of pathologic response to neoadjuvant therapy in breast cancer: Systematic review of the accuracy of MRI. Breast. 2012;21(5):669–677. | ||
Prevos R, Smidt ML, Tjan-Heijnen VCG, et al. Pre-treatment differences and early response monitoring of neoadjuvant chemotherapy in breast cancer patients using magnetic resonance imaging: a systematic review. Eur Radiol. 2012;22(12):2607–2616. | ||
Woolf DK, Padhani AR, Taylor NJ, et al. Assessing response in breast cancer with dynamic contrast-enhanced magnetic resonance imaging: Are signal intensity–time curves adequate? Breast Cancer Res Treat. 2014;147(2):335–343. | ||
Li SP, Makris A, Beresford MJ, et al. Use of dynamic contrast-enhanced MR imaging to predict survival in patients with primary breast cancer undergoing neoadjuvant chemotherapy. Radiology. 2011;260(1):68–78. | ||
Raunig DL, Mcshane LM, Pennello G, et al. Quantitative imaging biomarkers: A review of statistical methods for technical performance assessment. Stat Methods Med Res. 2015;24(1):27–67. | ||
Partridge SC, Mcdonald ES. Diffusion weighted magnetic resonance imaging of the breast: protocol optimization, interpretation, and clinical applications. Magn Reson Imaging Clin N Am. 2013;21(3):601–624. | ||
Park SH, Moon WK, Cho N, et al. Diffusion-weighted MR Imaging: Pretreatment Prediction of Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. Radiology. 2010;257(1):56–63. | ||
Belli P, Costantini M, Ierardi C, et al. Diffusion-weighted Imaging in Evaluating the Response to Neoadjuvant Breast Cancer Treatment. Breast J. 2011;17(6):610–619. | ||
Wu LM, Hu JN, Gu HY, Hua J, Chen J, Xu JR. Can diffusion-weighted MR imaging and contrast-enhanced MR imaging precisely evaluate and predict pathological response to neoadjuvant chemotherapy in patients with breast cancer? Breast Cancer Res Treat. 2012;135(1):17–28. | ||
Sardanelli F, Carbonaro LA, Montemezzi S, Cavedon C, Trimboli RM. Clinical Breast MR Using MRS or DWI: Who Is the Winner? Front Oncol. 2016;6(Suppl 1):217. | ||
Jagannathan NR, Kumar M, Seenu V, et al. Evaluation of total choline from in-vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br J Cancer. 2001;84(8):1016–1022. | ||
Meisamy S, Bolan PJ, Baker EH, et al. Neoadjuvant Chemotherapy of Locally Advanced Breast Cancer: Predicting Response with in Vivo 1H MR Spectroscopy—A Pilot Study at 4 T. Radiology. 2004;233(2):424–431. | ||
Baek HM, Chen JH, Nalcioglu O, Su MY, My S. Proton MR spectroscopy for monitoring early treatment response of breast cancer to neo-adjuvant chemotherapy. Ann Oncol. 2008;19(5):1022–1024. | ||
Yamaguchi K, Nakazono T, Egashira R, et al. Diagnostic Performance of Diffusion Tensor Imaging with Readout-segmented Echo-planar Imaging for Invasive Breast Cancer: Correlation of ADC and FA with Pathological Prognostic Markers. Magn Reson Med Sci. 2017;16(3):245–252. | ||
Furman-Haran E, Nissan N, Ricart-Selma V, Martinez-Rubio C, Degani H, Camps-Herrero J. Quantitative evaluation of breast cancer response to neoadjuvant chemotherapy by diffusion tensor imaging: Initial results. J Magn Reson Imaging. 2017. | ||
Fan M, Wu G, Cheng H, Zhang J, Shao G, Li L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur J Radiol. 2017;94:140–147. | ||
Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. | ||
Wang J, Kato F, Oyama-Manabe N, et al. Identifying Triple-Negative Breast Cancer Using Background Parenchymal Enhancement Heterogeneity on Dynamic Contrast-Enhanced MRI: A Pilot Radiomics Study. PLoS One. 2015;10(11):e0143308. | ||
Wu J, Gong G, Cui Y, Li R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging. 2016;44(5):1107–1115. | ||
Fan M, Cheng H, Zhang P, et al. DCE-MRI texture analysis with tumor subregion partitioning for predicting Ki-67 status of estrogen receptor-positive breast cancers. J Magn Reson Imaging. 2018;48(1):237–247. | ||
Ha S, Park S, Bang J-I, Kim E-K, Lee H-Y. Metabolic Radiomics for Pretreatment 18F-FDG PET/CT to Characterize Locally Advanced Breast Cancer: Histopathologic Characteristics, Response to Neoadjuvant Chemotherapy, and Prognosis. Sci Rep. 2017;7(1):1556. | ||
Park H, Lim Y, Ko ES, Es K, et al. Radiomics Signature on Magnetic Resonance Imaging: Association with Disease-Free Survival in Patients with Invasive Breast Cancer. Clinical Cancer Research. Epub 2018 Jun 18. | ||
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. | ||
Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18(3):e143–e152. | ||
Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: Evolving Considerations for PET response criteria in solid tumors. J Nucl Med. 2009;50 Suppl 1:122S–150. | ||
Weaver O, Leung JWT. Biomarkers and Imaging of Breast Cancer. AJR Am J Roentgenol. 2018;210(2):271–278. | ||
Li H, Zhu Y, Burnside ES, et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016;281(2):382–391. | ||
Pinker K, Shitano F, Sala E, et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging. 2018;47(3):604-620. | ||
Cx M, Gao F, Luo J, et al. NeoPalAna: Neoadjuvant Palbociclib, a Cyclin-Dependent Kinase 4/6 Inhibitor, and Anastrozole for Clinical Stage 2 or 3 Estrogen Receptor-Positive Breast Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23(15):4055–4065. | ||
Jovanović B, Mayer IA, Mayer EL, et al. A Randomized Phase II Neoadjuvant Study of Cisplatin, Paclitaxel With or Without Everolimus in Patients with Stage II/III Triple-Negative Breast Cancer (TNBC): Responses and Long-term Outcome Correlated with Increased Frequency of DNA Damage Response Gene Mutations, TNBC Subtype, AR Status, and Ki67. Clinical Cancer Research. 2017;23(15):4035–4045. | ||
Curigliano G, Romieu G, Campone M, et al. A phase I/II trial of the safety and clinical activity of a HER2-protein based immunotherapeutic for treating women with HER2-positive metastatic breast cancer. Breast Cancer Res Treat. 2016;156(2):301–310. | ||
Sturgeon C, Hill R, Hortin GL, Thompson D. Taking a new biomarker into routine use - A perspective from the routine clinical biochemistry laboratory. Proteomics Clin Appl. 2010;4(12):892–903. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.