Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 14
Prebiotic Colloidal Oat Supports the Growth of Cutaneous Commensal Bacteria Including S. epidermidis and Enhances the Production of Lactic Acid
Authors Liu-Walsh F, Tierney NK, Hauschild J, Rush AK , Masucci J, Leo GC, Capone KA
Received 28 August 2020
Accepted for publication 15 December 2020
Published 19 January 2021 Volume 2021:14 Pages 73—82
DOI https://doi.org/10.2147/CCID.S253386
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Fang Liu-Walsh,1 Neena K Tierney,1 James Hauschild,2 Allison K Rush,1 John Masucci,3 Gregory C Leo,3 Kimberly A Capone1
1Johnson & Johnson Consumer Inc., Skillman, NJ, USA; 2Johnson & Johnson Microbiological Quality & Sterility Assurance, Johnson & Johnson Inc., Raritan, NJ, USA; 3Janssen R&D Companies of Johnson & Johnson, Springhouse, PA, USA
Correspondence: Fang Liu-Walsh; Kimberly A Capone
Johnson & Johnson Consumer Inc., 199 Grandview Road, Skillman, NJ 08558-9418, USA
Email [email protected]; [email protected]
Background: Multiple skin conditions have been associated with alterations in the diversity and composition of the skin microbiome, including dry skin and atopic dermatitis. In these conditions, a number of commensal skin bacteria have been implicated in supporting a healthy skin barrier, including Staphylococcus epidermidis. Recent clinical studies in patients with mild-to-moderate atopic dermatitis and dry/itchy skin have shown significantly improved skin barrier function and microbial diversity upon treatment with moisturizers containing 1% colloidal oat. We hypothesized that direct use of colloidal oat by skin microbes contributes to these therapeutic benefits.
Methods: Skin bacterial growth was assessed using the BacT/ALERT system. Staphylococcus aureus and S. epidermidis growth rates and metabolism were compared in an in vitro competition assay. The effect of a 1% colloidal oat–containing moisturizer on lactic acid content of the stratum corneum was clinically assessed in subjects with moderate-to-severe dry skin. S. epidermidis gene expression was evaluated by next-generation mRNA sequencing. Short-chain fatty acids were quantified in bacterial culture supernatants.
Results: In vitro, colloidal oat increased the growth rate of S. epidermidis vs S. aureus, as well as the metabolism of S. epidermidis. Colloidal oat also significantly increased lactic acid concentrations in supernatants of both strains and decreased pH, consistent with clinical findings that 6-week use of a 1% colloidal oat–containing lotion significantly increased lactic acid on dry skin. Further analyses suggest that colloidal oat alters the gene expression profile of S. epidermidis.
Conclusion: Colloidal oat directly affects the growth, metabolism, lactic acid production, and gene expression of skin commensal bacteria, as shown via in vitro studies. The increased production of lactic acid reflects clinical observations with colloidal oat–containing skin moisturizers. Our findings suggest a new mechanism for colloidal oat as a skin prebiotic, which may contribute to improvements in skin and microbiome diversity in various skin conditions, including dry/itchy skin and atopic dermatitis.
Keywords: atopic dermatitis, dry skin, colloidal oat, skin barrier function, skin microbiome, skin moisturizer, S. epidermidis
Introduction
The skin is the largest organ of the body and home to a vast array of microbes.1 Phyla that dominate the skin are Actinobacteria, Proteobacteria, Bacteroidetes, and Firmicutes, with more than 60% of the bacterial species belonging to three genera, including Staphylococcus (Firmicutes); Corynebacterium (Actinobacteria), and Propionibacterium (Actinobacteria).2 Sites rich in sebaceous secretion are preferentially colonized by lipophilic bacteria such as Cutibacterium acnes (previously known as Propionibacterium acnes), whereas moist locations such as skin folds are favored by Corynebacterium spp. and Staphylococcus aureus. S. epidermidis is one of the most abundant bacterial species of the skin microbiome. It is a mutualistic, Gram-positive, facultative anaerobe that constitutes up to 90% of the aerobic resident flora, and certain strains have been associated with skin health.3
Multiple skin conditions have been associated with alterations in the diversity and composition of the skin microbiome, including atopic dermatitis (AD), dry skin, acne, dandruff/seborrheic dermatitis and psoriasis.4–6 For example, atopic dermatitis is commonly associated with colonization of S. aureus and a decrease in diversity.7–9 Interestingly, in contrast, dry skin is associated with an increase in microbial diversity along with a decrease in microbial load in comparison to more sebaceous areas of the skin.6 Acne presents as an excessive growth of pathogenic C. acnes,10,11 and in dandruff/seborrheic dermatitis there is an uncontrolled growth of Malassezia species.12,13 While these examples provide ample illustration of the association between skin disease/skin condition and alterations in microbiome composition, the precise role of alterations in these skin states and the events that trigger these alterations are not well understood.
Understanding the overall composition of the skin barrier and how these components of the skin influence or are influenced by the skin microbiota is also pivotal to identifying the role and benefits of the microbiome in the health of the skin.
Lactic acid is one of the natural moisturizing factors (NMF) of the stratum corneum skin barrier that helps to maintain the hydration and slightly acidic pH (ie, “acid mantle”) of the skin surface.14–17 Among the NMF components, lactic acid and potassium concentrations are the only ones that have been shown to significantly correlate with the level of skin hydration and skin surface pH (lower lactic acid in dry skin and skin with elevated pH).15 In this regard, higher lactic acid concentrations and lower skin surface pH have been associated with increased epidermal renewal in healthy skin subjects,14 and potassium lactate application has been shown to improve skin surface hydration in patients with mild AD.17 Although it is well accepted that the presence of lactic acid in the skin is crucial for optimal skin hydration, the precise source of lactic acid on the skin is not well defined.
Prebiotics are a well-established concept in digestive health. They are non-digestible food ingredients that selectively stimulate the growth of beneficial species (mainly bifidobacteria and lactobacilli) of the intestinal microbiota.18 These bacteria use prebiotics as substrates to produce metabolites and micronutrients that may benefit the host.19 Although there is no established concept for prebiotics for the skin, we suggest that a topically applied ingredient that is metabolized by members of the skin microbiome and promotes changes in the composition and/or activity of the skin microbiome to benefit the skin may be considered a prebiotic for the skin.20
Oat bran or oat fiber and their main active ingredient, β-glucans, have been well documented to serve as prebiotics in the gut.19,21–24 Oat (Avena sativa) in colloidal form has been used for centuries as a topical treatment of skin conditions such as AD and other dry, compromised skin conditions.25,26 Additionally, in recent clinical studies in patients with mild-to-moderate AD or dry/itchy skin, 2 weeks of treatment with moisturizers containing 1% colloidal oat significantly improved skin barrier properties, including decreasing transepidermal water loss and increasing skin hydration, as well as increasing microbial diversity.27 We hypothesized that the therapeutic benefit of these moisturizers may be related, at least in part, to the prebiotic properties of colloidal oat, which contains 65% to 85% starch, 15% to 20% protein, 3% to 11% lipids, 5% fiber, and 5% β-glucans, as a substrate.25,28 To test this hypothesis, we examined the effects of 1% colloidal oat on the growth rate, metabolism, lactic acid production, and gene expression of S. epidermidis and S. aureus via in vitro studies. Additionally, we clinically evaluated the lactic acid content in the skin barrier after topical application of a 1% colloidal oatmeal–containing moisturizer on subjects with moderate-to-severe dry skin.
Materials and Methods
Bacterial Strains
Bacterial strains were obtained from the American Type Culture Collection (ATCC), including S. epidermidis (ATCC 12,228), S. aureus (ATCC 6538), and C. acnes (ATCC 6919).
Bacterial Growth: Time-to-Detection (TTD) by BacT/ALERT Based on CO2 Detection
The BacT/ALERT system, which is used extensively in hospital clinical laboratories for the detection of microorganisms in blood samples,29–33 is a fully automated microbial detection system based on the colorimetric detection of CO2 in sample bottle growth media. A BacT/ALERT prebiotic screening method was developed based on the relative TTD of target microorganism in the absence and presence of potential prebiotic ingredients. In this study, the relative TTD of target microorganisms with and without the addition of 1% colloidal oat was evaluated. The TTD of target microorganisms in the absence of 1% colloidal oat was set at 100% (control), and the relative TTD in the presence of 1% colloidal oat was expressed as a percent of the control. The presence of a prebiotic effect results in a shorter TTD compared with the control and is reported as a relative TTD below 100%.
The target microorganisms to be tested were harvested from BacT/ALERT sample bottles containing overnight stationary phase cultures, which were serially diluted into new BacT/ALERT sample bottles. One-mL aliquots containing approximately 1000 colony-forming units (CFU) were inoculated into designated BacT/ALERT sample bottles using a sterile 1-mL syringe and 20-gauge needle. Prior to microbial inoculation, 10-mL aliquots of oat suspension were aseptically injected into designated sample bottles using a sterile 10-mL syringe and 18-gauge needle, to obtain a 1% final concentration of colloidal oat. The oat suspension contained 0.51 g resuspended oat per 10 mL volume of sterile filtered water for injection (WFI)-quality water. Control sample bottles inoculated with the designated test microorganisms contained only WFI in the absence of 1% colloidal oat. A negative control containing 1% colloidal oat only was included.
In vitro Bacterial Competition Assay
Co-cultures of S. epidermidis and S. aureus were inoculated at 1000 CFU/mL in 125-mL sterile flasks containing 50 mL of Tryptic Soy Broth (TSB: 1% tryptone [Difco], 0.5% yeast extract [Gibco-BRL], 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) with or without 1% colloidal oat. Flasks were incubated at 33°C ± 2°C, and samples were taken at 0, 2, 4, 6, and 24 h. Serial dilutions of each sample were plated in duplicate on Mannitol Salt Agar plates, and colonies were counted after incubation at 33°C ± 2°C for 48 h. The use of Mannitol Salt Agar allowed for the differentiation of S. aureus and S. epidermidis on the co-cultured competition assay plates. Mannitol fermentors such as S. aureus appeared as yellow colonies with yellow zones in the media while the non-mannitol fermentor, S. epidermidis, appeared as clear pink to red colonies with no yellow color change in the medium. For each culture condition and time point, titers were determined from five to eight replicate samples. Non-parametric, two-sided Mann–Whitney tests were used to assess the statistical significance of differences in bacterial concentrations between culture conditions.
Analysis of Short-Chain Fatty Acids
Analysis and quantification of short-chain fatty acids (SCFAs) were performed by gas chromatography followed by mass spectrometry using a 7890A gas chromatograph and a 5975C mass spectrometer (Agilent Technologies). Quantification of lactic acid and succinic acid was performed by high-pressure liquid chromatography (Shimadzu LC-20A System) followed by tandem mass spectrometry (AB Sciex API 4000 with TurboIonSpray® Source). All analyses were performed in triplicate. Unsupervised hierarchical clustering analysis was used to compare metabolic activity (ie, SCFA production) of S. epidermidis under different culture conditions.
Gene Expression Analyses
High-throughput RNA sequence (RNA-seq) reads were mapped to the S. epidermidis genome using the Rockhopper pipeline.34 A negative binomial model (edgeR) was used to evaluate differential gene expression, with read counts normalized using the weighted trimmed mean of M-values method, and dispersions estimated using the Cox-Reid–adjusted likelihood approach. Differential expression analyses were performed using features with detectable expression in at least two of six samples involved in a given comparison. A feature was considered to have detectable expression in a sample if the count per million mapped reads estimate was greater than 0.20. The StringDB database was used for the functional analysis of differentially expressed genes.35
Clinical Study of Colloidal Oat-Containing Daily Moisturizer in Subjects with Dry Skin
A single-center clinical study was conducted on 18- to 65-year-old female subjects with Fitzpatrick skin type I–IV who had clinically determined moderate-to-severe dry skin on the lower leg and used moisturizer at least one to three times per week (registered at Clinicaltrials.gov NCT04510103). One objective of the study was to determine the impact of a daily moisturizing lotion containing 1% colloidal oat on the amount of lactic acid, a natural moisturizing factor, within the stratum corneum skin barrier. The lotion was applied twice a day for 6 weeks after a 3-day washout period with a gentle skin cleanser that was continued for the study duration. At baseline and after 6-weeks use of the lotion, two consecutive D-Squame Standard Sampling Discs (CuDerm Corporation, Dallas, TX) were collected from the designated test site. The first tape, collected from the uppermost layer of the stratum corneum, was used to quantify the degree of skin flakiness by image analysis. The lactic acid content in the second tape was extracted, quantitated via colorimetry, and normalized by total protein content (Synelvia SAS, Labège, France). A paired t-test was performed to compare the lactic acid content at baseline and after 6 weeks of treatment for six subjects. The trial was conducted in accordance with the Declaration of Helsinki. Each participant provided informed consent. The IntegReview Institutional Review Board (Austin, TX) approved the study.
Results
Differential Effects of 1% Colloidal Oat on the Growth of Members of the Skin Microbiome
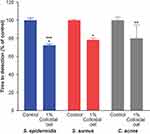
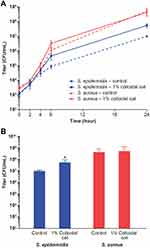
Using the BacT/Alert system for detection of S. epidermidis, S. aureus, and C. acnes grown in bacterial monocultures, we found that addition of 1% colloidal oat to the culture medium significantly reduced the TTD compared with cultures grown in the absence of 1% colloidal oat (control) (Figure 1). On average, TTD of S. epidermidis, S. aureus, and C. acnes grown with 1% colloidal oat was 72%, 78%, and 80% that of the corresponding time in control cultures. Importantly, the increase in growth rate obtained with 1% colloidal oat was significantly greater for S. epidermidis than S. aureus (p<0.001). This suggests that the beneficial effects of moisturizers containing 1% colloidal oat observed in patients with AD or dry/itchy skin28,36 may be attributable, at least in part, to the effects of 1% colloidal oat on the increase in diversity of the microbiome overall, along with the increase in the relative abundance of S. aureus and S. epidermidis species. To explore this possibility further, we tested the effect of 1% colloidal oat on the growth of S. aureus and S. epidermidis in an in vitro competition assay. In the absence of oat (control), S. aureus grew faster than S. epidermidis (Figure 2A and B). However, although the addition of 1% colloidal oat to the culture medium had no substantial effect on the growth of S. aureus (mean titer after 24 h, 4 × 108 CFU/mL vs 5 × 108 CFU/mL without oat), it selectively increased the growth rate of S. epidermidis (mean titer after 24 h, 5 × 107 CFU/mL vs 1 × 107 CFU/mL without oat; p=0.012, two-tailed Mann–Whitney test; Figure 2A and B).
Effects of 1% Colloidal Oat on S. epidermidis Metabolism and Lactic Acid Production
Metabolism of 1% colloidal oat in mixed cultures of S. aureus and S. epidermidis (1:1 ratio) resulted in significant decreases in the pH of the culture media from a mean value ± standard deviation of 7.61 ± 0.29 to 7.15 ± 0.38; p<0.001.
A previous study showed that fermentation of oat by fecal bacterial generated significantly more butyric acid than fermentation of other grains, such as corn and wheat.37 Interestingly, in our study fermentation of 1% colloidal oat by the tested skin bacteria did not yield detectable levels of butyric acid (Figure 3A). Propionic acid was generated only by C. acnes (among the tested bacterial strains), and its concentration was not significantly affected by the presence of 1% colloidal oat (1459.8 ± 35.4 μg/mL without 1% colloidal oat and 1470.5 ± 4.5 μg/mL with 1% colloidal oat). Under aerobic conditions, 1% colloidal oat altered the metabolite composition of both S. aureus and S. epidermidis, with significantly increased production of lactic acid by factors of 1.7 (from 913.4 ± 30.0 μg/mL to 1581.5 ± 66.7 µg/mL, p=0.0001) and 3.1 (from 687.2 ± 134.4 μg/mL to 2137.7 ± 92.2 μg/mL, p=0.0082), respectively (Figure 3A and B). The levels of lactic acid produced by S. epidermidis in the presence of 1% colloidal oat were particularly high and very similar to those produced by Lactococcus lactis (used as a positive control for lactic acid production) in the presence or absence of colloidal oat. The highest levels of lactic acid were produced by S. epidermidis under anaerobic conditions (Figure 3A). Unsupervised hierarchical clustering analysis further showed that the metabolic activity of S. epidermidis under aerobic conditions and in the presence of 1% colloidal oat is more similar to that under anaerobic conditions than that under aerobic conditions in the absence of 1% colloidal oat (Figure 4).
 |
Figure 4 Unsupervised hierarchical clustering analysis of S. epidermidis metabolic activity under different culture conditions. |
To support the results of the in vitro studies, clinical evaluations with a moisturizer containing 1% colloidal oat demonstrated the mean lactic acid content in vivo was significantly increased (by a factor of 1.9; p<0.05) in moderate-to-severe dry skin after 6 weeks of twice-daily application (Figure 5). This suggests that 1% colloidal oat might contribute to the production of SCFAs, including lactic acid, by various skin bacteria.
Effects of 1% Colloidal Oat on S. epidermidis Gene Expression
The gene expression profiles of S. epidermidis treated with and without 1% colloidal oat were evaluated using RNA-seq. S. epidermidis was grown in TSB media under aerobic conditions at 33°C. Overall, 418 genes were significantly upregulated ≥2-fold, and 335 genes were significantly downregulated ≥2-fold when treated with 1% colloidal oat (Supplementary Materials). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed with genes significantly upregulated or downregulated to dissect the molecular pathways affected by treatment with 1% colloidal oat. Significantly enriched KEGG pathways and associated genes that were differentially expressed with versus without 1% colloidal oat are shown in Table 1.
 |
Table 1 Enriched KEGG Pathways of S. epidermidis Differential Gene Expression in the Presence of 1% Oat |
The top enriched KEGG pathway of genes significantly downregulated was the glycolysis pathway; six of 10 key enzymes in the glycolysis pathway were significantly decreased when 1% colloidal oat was present in the medium, suggesting a shift in bacterial metabolism from simple sugars in the culture medium to the complex carbohydrates in oat. Genes significantly upregulated in the presence of 1% colloidal oat included genes important for cell proliferation, DNA synthesis, and adenosine triphosphate (ATP) metabolism, consistent with the observed increased growth of the S. epidermidis in the presence of oat. In addition, alpha-D-1,4-glucosidase and sucrose-6-phosphate hydrolase, enzymes involved in breaking down starch, were significantly upregulated. KEGG pathway analysis further revealed that bacterial infection pathway were enriched significantly, including the genes dltA, B, C and D, which are part of the same operon.
Discussion
Our findings demonstrate that 1% colloidal oat is used by skin bacteria and influences their growth rate, metabolism, production of lactic acid, and gene expression. Specifically, in an in vitro competition assay, 1% colloidal oat selectively increased the growth rate of S. epidermidis, a commensal skin resident associated with skin health, versus S. aureus.
Our in vitro data suggest that skin bacteria, including S. epidermidis, may be one of the major sources of lactic acid, and that metabolism of 1% colloidal oat increases lactic acid production by skin bacteria. The observed increase in lactic acid production with 1% colloidal oat was greater with S. epidermidis, a commensal bacterium, than with S. aureus, an opportunistic pathogen. Clinical evaluation with a 1% colloidal oat–containing moisturizer supported these in vitro results, demonstrating an increase in the lactic acid content in the skin barrier. Additionally, our in vitro results showed that the bacterial strains tested do not produce major amounts of butyric acids, which is beneficial for the gut but can be harmful to the skin.38,39 Propionic acid was generated only by C. acnes, and 1% colloidal oat had no significant influence on its production.
Under anaerobic conditions, S. epidermidis grows faster than S. aureus (data not shown). Comparison of metabolite profiles including hierarchical clustering analysis revealed that S. epidermidis grown under aerobic conditions in the presence of 1% colloidal oat displayed a metabolic profile more similar to that under anaerobic conditions than to that under aerobic conditions in the absence of oat, suggesting a shift in metabolic pathways.
Analysis of differential gene expression revealed that the addition of 1% colloidal oat under aerobic conditions rendered large changes in the gene expression profile of the S. epidermidis. Gene expression profiling through mRNA sequencing identified 2665 genes expressed in S. epidermidis. Among those, 418 genes were significantly upregulated and 335 genes significantly downregulated upon 1% colloidal oat treatment.
As expected, oat significantly changed the metabolism of the S. epidermidis. Of note, six of 10 key enzymes in the glycolysis pathway were decreased significantly. The TSB medium S. epidermidis was grown in a nutrient-rich medium with high concentrations of simple sugars such as glucose. Our findings suggest that in the presence of 1% colloidal oat in the medium, S. epidermidis relied less on glycolysis of simple sugars and more on metabolism of the complex carbohydrates in colloidal oat. Consistent with this interpretation, alpha-D-1,4-glucosidase and sucrose-s-phosphate hydrolase, enzymes involved in breaking down starch (a major ingredient of colloidal oat) were significantly increased in the presence of 1% colloidal oat. Consistent with the growth assay, genes related to bacterial growth, such as those involved in DNA synthesis, amino acid synthesis, and ATP metabolism, were upregulated when 1% colloidal oat was present in the growth medium.
KEGG pathway analysis revealed that dltA, dltB, dltC and dltD, which are from the same operon, were co-upregulated. This group of genes has been shown to be involved in the metabolism of D-alanine, a component of bacterial cell wall peptidoglycan. Interestingly, dltA has been shown to help bacteria to persist on human skin through evasion of cutaneous innate defense by cationic skin-derived antimicrobial peptides.40 Thus, dltA may help S. epidermidis to outcompete opportunistic skin pathogens, such as S. aureus.
In summary, taken together these findings suggest direct effects of 1% colloidal oat on the growth rate, metabolism, lactic acid production, and gene expression of commensal skin microbes. These effects may contribute to the therapeutic benefits and increased microbial diversity observed in various skin conditions, such as dry/itchy skin and atopic dermatitis, upon application of 1% colloidal oatmeal-containing skin moisturizers.
Data Sharing Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Compliance with Ethical Standards
Some of the research described in this report involved a study in human participants. The trial was conducted in accordance with the Declaration of Helsinki. The study was approved by IntegReview Institutional Review Board (Austin, TX). Each participant provided informed consent.
Acknowledgments
The authors thank Ann Ballesteros, PhD, and Karen Meyer, for her support in the clinical study. Bacterial RNA extraction for RNA sequencing was performed by Sunny BioDiscovery, Inc., and statistical analysis of RNA sequence reads was performed by William Swindell of Ohio University Heritage College of Osteopathic Medicine, with input from Jeremy Ranfaing, PhD, of Johnson and Johnson. Writing assistance was provided by Roland Tacke, PhD, of Evidence Scientific Solutions Inc, funded by Johnson & Johnson Consumer Inc.
Disclosures
All authors are employees and own stock of Johnson & Johnson. The authors report no other potential conflicts of interest for this work.
Funding
This study was funded by Johnson & Johnson Consumer Inc.
References
1. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi:10.1038/nrmicro2537
2. Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi:10.1126/science.1171700
3. Baviera G, Leoni MC, Capra L, et al. Microbiota in healthy skin and in atopic eczema. Biomed Res Int. 2014;2014:436921. doi:10.1155/2014/436921
4. Egert M, Simmering R. The microbiota of the human skin. Adv Exp Med Biol. 2016;902:61–81.
5. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi:10.1038/nature18848
6. Grice EA. The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin Cutan Med Surg. 2014;33(2):98–103. doi:10.12788/j.sder.0087
7. Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi:10.1101/gr.131029.111
8. Williams MR, Nakatsuji T, Sanford JA, Vrbanac AF, Gallo RL. Staphylococcus aureus induces increased serine protease activity in keratinocytes. J Invest Dermatol. 2017;137(2):377–384. doi:10.1016/j.jid.2016.10.008
9. Harkins CP, Pettigrew KA, Oravcová K, et al. The microevolution and epidemiology of Staphylococcus aureus colonization during atopic eczema disease flare. J Invest Dermatol. 2018;138(2):336–343. doi:10.1016/j.jid.2017.09.023
10. Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep. 2016;6(1):39491. doi:10.1038/srep39491
11. Ryu S, Han HM, Song PI, Armstrong CA, Park Y. Suppression of Propionibacterium acnes infection and the associated inflammatory response by the antimicrobial peptide P5 in mice. PLoS One. 2015;10(7):e0132619. doi:10.1371/journal.pone.0132619
12. Loing E, Lamarque E, Borel M. New targets in the battle against dandruff. J Cosmet Sci. 2017;68(1):107–113.
13. Borda LJ, Wikramanayake TC. Seborrheic dermatitis and dandruff: a comprehensive review. J Clin Investig Dermatol. 2015;3(2):
14. Thueson DO, Chan EK, Oechsli LM, Hahn GS. The roles of pH and concentration in lactic acid-induced stimulation of epidermal turnover. Dermatol Surg. 1998;24(6):641–645. doi:10.1111/j.1524-4725.1998.tb04221.x
15. Nakagawa N, Sakai S, Matsumoto M, et al. Relationship between NMF (lactate and potassium) content and the physical properties of the stratum corneum in healthy subjects. J Invest Dermatol. 2004;122(3):755–763. doi:10.1111/j.0022-202X.2004.22317.x
16. Sharquie KE, Al-Tikreety MM, Al-Mashhadani SA. Lactic acid as a new therapeutic peeling agent in melasma. Dermatol Surg. 2005;31(2):149–154. doi:10.1097/00042728-200502000-00005
17. Sugawara T, Kikuchi K, Tagami H, Aiba S, Sakai S. Decreased lactate and potassium levels in natural moisturizing factor from the stratum corneum of mild atopic dermatitis patients are involved with the reduced hydration state. J Dermatol Sci. 2012;66(2):154–159. doi:10.1016/j.jdermsci.2012.02.011
18. Roberfroid M, Gibson GR, Hoyles L, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010;104(Suppl 2):S1–S63. doi:10.1017/S0007114510003363
19. Kedia G, Vázquez JA, Charalampopoulos D, Pandiella SS. In vitro fermentation of oat bran obtained by debranning with a mixed culture of human fecal bacteria. Curr Microbiol. 2009;58(4):338–342. doi:10.1007/s00284-008-9335-1
20. Lanzalaco AC, Charbonneau DL, Howard BW, inventors. Patent: Topical use of a skin-commensal prebiotic agent and compositions containing the same. International patent WO 2013/122932 A2. 2013 Aug 22.
21. Connolly ML, Tuohy KM, Lovegrove JA. Wholegrain oat-based cereals have prebiotic potential and low glycaemic index. Br J Nutr. 2012;108(12):2198–2206. doi:10.1017/S0007114512000281
22. Coman MM, Verdenelli MC, Cecchini C, et al. Effect of buckwheat flour and oat bran on growth and cell viability of the probiotic strains Lactobacillus rhamnosus IMC 501®, Lactobacillus paracasei IMC 502® and their combination SYNBIO®, in synbiotic fermented milk. Int J Food Microbiol. 2013;167(2):261–268. doi:10.1016/j.ijfoodmicro.2013.09.015
23. Zhang N, Li D, Zhang X, Shi Y, Wang H. Solid-state fermentation of whole oats to yield a synbiotic food rich in lactic acid bacteria and prebiotics. Food Funct. 2015;6(8):2620–2625. doi:10.1039/C5FO00411J
24. Carlson JL, Erickson JM, Hess JM, Gould TJ, Slavin JL. Prebiotic dietary fiber and gut health: comparing the in vitro fermentations of beta-glucan, inulin and xylooligosaccharide. Nutrients. 2017;9(12):1361. doi:10.3390/nu9121361
25. Cerio R, Dohil M, Jeanine D, Magina S, Mahé E, Stratigos AJ. Mechanism of action and clinical benefits of colloidal oatmeal for dermatologic practice. J Drugs Dermatol. 2010;9(9):1116–1120.
26. Reynertson KA, Garay M, Nebus J, et al. Anti-inflammatory activities of colloidal oatmeal (Avena sativa) contribute to the effectiveness of oats in treatment of itch associated with dry, irritated skin. J Drugs Dermatol. 2015;14(1):43–48.
27. Capone K, Klein SL, Kirchner F, Tierney NK. Effects of topical lotions on the atopic dermatitis skin microbiome and associations with itch and skin barrier function.
28. Futterer TJ, Tierney NK, Rush A, Meyer K, Capone K. Assessment of skin microbiome diversity and skin health in dry skin and dry, itchy skin: a bilateral, controlled clinical trial using oat-containing lotions and wash. J Am Acad Dermatol. 2019;81(4 Suppl 1):AB23.
29. Jacobs MR, Mazzulli T, Hazen KC, et al. Multicenter clinical evaluation of BacT/Alert Virtuo blood culture system. J Clin Microbiol. 2017;55(8):2413–2421. doi:10.1128/JCM.00307-17
30. Nutman A, Fisher Even-Tsur S, Shapiro G, Braun T, Schwartz D, Carmeli Y. Time to detection with BacT/Alert FA plus compared to BacT/Alert FA blood culture media. Eur J Clin Microbiol Infect Dis. 2016;35(9):1469–1473. doi:10.1007/s10096-016-2686-9
31. Bugno A, Lira RS, Oliveira WA, Almodovar AA, Saes DP, Pinto TJ. Application of the BacT/ALERTR 3D system for sterility testing of injectable products. Braz J Microbiol. 2015;46(3):743–747. doi:10.1590/S1517-838246320140587
32. Bourbeau P, Riley J, Heiter BJ, Master R, Young C, Pierson C. Use of the BacT/Alert blood culture system for culture of sterile body fluids other than blood. J Clin Microbiol. 1998;36(11):3273–3277. doi:10.1128/JCM.36.11.3273-3277.1998
33. Ziegler R, Johnscher I, Martus P, Lenhardt D, Just HM. Controlled clinical laboratory comparison of two supplemented aerobic and anaerobic media used in automated blood culture systems to detect bloodstream infections. J Clin Microbiol. 1998;36(3):657–661. doi:10.1128/JCM.36.3.657-661.1998
34. McClure R, Balasubramanian D, Sun Y, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41(14):e140. doi:10.1093/nar/gkt444
35. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–D452. doi:10.1093/nar/gku1003
36. Capone K, Kirchner F, Klein SL, Tierney NK. Effects of colloidal oatmeal topical atopic dermatitis cream on skin microbiome and skin barrier properties. J Drugs Dermatol. 2020;19(5):524–531. doi:10.36849/JDD.2020.4924
37. Casterline JL, Oles CJ, Ku Y. In vitro fermentation of various food fiber fractions. J Agric Food Chem. 1997;45(7):2463–2467. doi:10.1021/jf960846f
38. Ohira H, Tsutsui W, Fujioka Y. Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis. J Atheroscler Thromb. 2017;24(7):660–672. doi:10.5551/jat.RV17006
39. Sanford JA, Zhang LJ, Williams MR, Gangoiti JA, Huang CM, Gallo RL. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol. 2016;1(4):eaah4609. doi:10.1126/sciimmunol.aah4609
40. Simanski M, Glaser R, Köten B, et al. Staphylococcus aureus subverts cutaneous defense by D-alanylation of teichoic acids. Exp Dermatol. 2013;22(4):294–296. doi:10.1111/exd.12114
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.