Back to Journals » OncoTargets and Therapy » Volume 13
PRDX1 is a Tumor Suppressor for Nasopharyngeal Carcinoma by Inhibiting PI3K/AKT/TRAF1 Signaling
Authors Xiao H, Yang T, Yan L, Feng J, Huang B, Jiang Y
Received 3 March 2020
Accepted for publication 15 July 2020
Published 15 September 2020 Volume 2020:13 Pages 9123—9133
DOI https://doi.org/10.2147/OTT.S252286
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Hongmei Xiao,1,2 Taoyu Yang,3 Lingli Yan,4 Jihong Feng,5 Boyan Huang,6 Yu Jiang1
1Department of Medical Oncology, Cancer Center, West China Hospital, Sichuan University, Chengdu 610041, People’s Republic of China; 2Oncology Department, Affiliated Hospital of Zunyi Medical University, Zunyi, People’s Republic of China; 3Department of Invasive Technology, The Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan 511500, People’s Republic of China; 4Department of Immunology, Medical University, Zunyi 563000, Guizhou, People’s Republic of China; 5Department of Oncology, Taizhou City People’s Hospital, Taizhou 318000, People’s Republic of China; 6National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen 518000, People’s Republic of China
Correspondence: Yu Jiang
Department of Medical Oncology, Cancer Center, West China Hospital, Sichuan University, Chengdu 610041, People’s Republic of China
Email [email protected]
Background: Peroxiredoxin 1 (PRDX1) has been identified as a dual regulator of tumorigenesis. However, its expression, clinical significance, and biological function in nasopharyngeal carcinoma (NPC) remain unknown. This study aimed to explore the role and underlying mechanisms of PRDX1 in NPC.
Materials and Methods: The expression of PRDX1 in NPC tissues was evaluated by immunohistochemistry, and the relationships between the expression of PRDX1 and clinical features and prognosis of NPC patients were analyzed. The effects of PRDX1 on NPC cell proliferation, migration, invasion, and epithelial-to-mesenchymal transition (EMT) were examined. A tumor-bearing model of nude mouse was established to verify the function of PRDX1 in vivo.
Results: PRDX1 expression level was negatively associated with recurrence and metastasis of NPC. PRDX1 knockdown promoted NPC cell proliferation, migration, invasion and EMT in vitro, and enhanced tumor growth in vivo, while PRDX1 overexpression had opposite effects. Furthermore, transcriptome analysis showed that PRDX1 inhibited the activation of PI3K/AKT/TRAF1 signaling in NPC cells.
Conclusion: PRDX1 inhibits NPC by inhibiting the activation of PI3K/AKT/TRAF1 signaling. PRDX1 is a tumor suppressor in human NPC and may be a prognostic biomarker for NPC patients.
Keywords: PRDX1, NPC, proliferation, migration, invasion, EMT, PI3K/AKT, TRAF1
Introduction
Human nasopharyngeal carcinoma (NPC) is a common malignant tumor of the head and neck.1 NPC shows distinct regional and ethnic differences in epidemiologic features.2,4 Although the prognosis of NPC patients is improved due to the advancement of chemotherapy and radiation therapy, distant metastasis is the main reason for the failure of treatment failure or even death.5
The association between reactive oxygen species (ROS) and tumor environment in cancer progression has been well illustrated. Peroxiredoxin 1 (PRDX1) is ubiquitously expressed in eukaryotic cells and participates in oxidization-reduction balance and peroxide detoxification via eliminating ROS.6 PRDX1 plays a role in cell proliferation, tumor promotion, and apoptotic processes in many human cancers.7,10 Cai et al11 found that PRDX1 could promote esophageal squamous cell carcinoma metastasis by regulating the AKT/mTOR pathway. Overexpression of PRDX1 could enhance TGF-β1-induced epithelial-to-mesenchymal transition (EMT) and cell migration.12 However, reduction of PRDX1 could activate the PI3K/AKT pathway to promote breast cancer.13 In addition, silencing of PRDX1 downregulated VEGF expression in prostate cancer cells by activating TLR4 and NF-KB.14 Taken together, PRDX1 seems to have the dual role of promoting or inhibiting cancers. Up to now, the role of PRDX1 in NPC remains unclear.
In this study, we found that PRDX1 was expressed at a low level in human NPC tissues. Furthermore, we revealed that PRDX1 inhibited NPC cell proliferation and EMT, mainly depending on the PI3K/AKT/TRAF1 signal pathway.
Materials and Methods
Tissue Samples
This study was approved by the Medical Ethical Committee of the Affiliated Hospital of Zunyi Medical University, and all patients provided written informed consent. A total of 89 patients with NPC not treated by radiotherapy and chemotherapy before were enrolled from January 2004 to December 2016. All the tumor samples were diagnosed by two qualified pathologists and the clinical staging was defined by two clinicians.
Immunohistochemistry
The immunohistochemistry (IHC) method was used to analyze PRDX1 expression in paraffin sections fixed in paraformaldehyde using the protocol as described previously.15 Immunohistochemical scores were obtained based on staining intensity and the percentage of positively stained cells.15
Cell Culture
The NPC cell lines CNE1, CNE2, 6–10B, and C666-1 and human nasopharyngeal epithelial cell line NP69 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, USA) and penicillin–streptomycin in a humidified incubator at 37ºC with 5% CO2.
Lentiviral Vectors
The lentivirus harboring sh-PRDX1 or PRDX1 cDNA and control vectors were purchased from Jikai (Shanghai, China). Cells were infected with lentivirus at the multiplicity of infection (MOI) of 80, and the cells were cultured in medium with 2 μg/mL puromycin (Servicebio, China) for 1 week, and then cultured in medium with 1 μg/mL puromycin for 7 days to obtain stable cell lines with overexpression or knockdown of PRDX1. The names and sequences of the vectors were shown in Table 1.
 |
Table 1 Sequences of All siRNAs Used in This Study |
QRT-PCR Analysis
Total RNA was extracted using TRIZOL (TaKaRa, Japan) and was reverse transcribed into cDNA. PCR was performed by using SYBR Premix Ex Taq (TaKaRa). Amplification conditions were preheating at 95°C (30 seconds), amplified at 95°C (5 seconds) and 60°C (30 seconds) for 40 cycles. The data of qRT-PCR were calculated using the 2-ΔΔCt method. The primer sequences are listed in Table 2.
 |
Table 2 Sequences of Primers Used in This Study |
Western Blot Analysis
Western blot analysis was performed using a routine protocol as described previously.16 Antibodies used were against PRDX1 (OriGene, USA), E-cadherin, Vimentin, N-cadherin, snail, twist1 (all Abcam, UK), AKT, p-AKT, PI3K, p-PI3K, TRAF1 (all Cell Signaling Technology, USA), p65, p-p65, p53 (all Abcam), Ki67 (OriGene, USA), and GAPDH (Abcam).
Cell Proliferation Assay
The cells were seeded in 96-well plates and cultured for 4 days. The CCK-8 solution was then added according to instruction of the CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan). After 2 hours incubation, the absorbance at 450 nm was measured using a microplate reader (BIORAD, iMark Microplate Reader, USA).
Scratch Wound Healing Assay
The cells were seeded in 6-well plates at 4×105/well per well. When the cells were completely confluent at the bottom wall, three scratches were made in each well with a micro-tip. Cells were rinsed gently twice with PBS to remove the detached cells. The plates were placed in an incubator for 24 hours and photographed under a microscope.
Migration and Invasion Assays
For migration assay, cells were prepared as a suspension of 1×105 cells/mL in serum-free medium. A 24-well culture plate was used as the lower chamber, and a transwell chamber was used as the upper chamber. A total of 600 µL of culture medium containing 20% fetal bovine serum was added to each well of the lower chamber, and 100 µL of the cell suspension was added to the upper chamber. The culture plate was placed in an incubator for 24 hours, and then the lower chamber was washed. The chamber was immersed in paraformaldehyde for 30 minutes at room temperature and rinsed. After staining with 0.1% crystal violet for 10 minutes and rinsing, photographs were taken under a microscope. The number of cells was counted in three random fields, and the average number of migrated cells was calculated. Invasion assay was basically the same as migration assay except the chamber was percolated with matrigel.
Animal Xenograft
Animal experiments were approved by the Animal Use and Care Committee of Sichuan University with strict accordance of the Chinese National Guidelines for Experimental Animal Welfare. 5-week old BALB/c nude mice (weight 16–18 g) were purchased from the Animal Laboratory Center of West China Hospital (China). PRDX1 overexpression or knockdown cell lines and control cells were inoculated subcutaneously under the right axilla of nude mice, and the tumor size was measured every 3 days.
Transcriptome Sequencing
Total RNA was isolated from CNE2 cells using the Trizol Reagent (Invitrogen), and the quality and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific). For mRNA sequencing, a Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA) and random oligonucleotides and SuperScript III were used to synthesize the first strand cDNA. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities and the enzymes were removed. After adenylation of the 3′ ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. To select cDNA fragments of the preferred 300 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, CA, USA). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using an Illumina PCR Primer Cocktail in a 15 cycle PCR reaction. Products were purified (AMPure XP system) and quantified using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent). The library was then sequenced on a NextSeq 500 platform (Illumina) by Shanghai Personal Biotech.
Statistical Analysis
Statistical analysis was performed using the SPSS 22.0 software package and GraphPad Prism 7.0. All experimental results were expressed as the mean±standard deviation (SD). Comparisons were performed using variance analysis. The clinical characteristics and expression of PRDX1 were evaluated by χ2 test. Survival analysis was performed using the Kaplan-Meier method and Log rank test. P<0.05 was considered statistically significant.
Results
PRDX1 Expression is Decreased in NPC Tissues
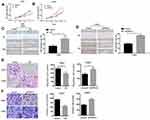
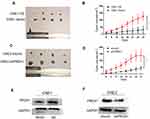
According to univariate analysis, we evaluated the association between PRDX1 expression and clinicopathological characteristics (Table 3). PRDX1 expression had no significant association with gender, age, tumor size, histological grade, and clinical stage (all P>0.05). Kaplan-Meier survival curves showed that both local recurrence rate (Figure 1A) and distant metastasis rate (Figure 1B) in patients with PRDX1 positive expression were less than the control group (P=0.037 and P=0.04, respectively). Typical IHC staining of PRDX1 in NPC tissues is shown in Figure 1C. PRDX1 positive expression (staining scores >3) was 34.83% (31/89). These data suggest that PRDX1 may be a tumor suppressor in NPC.
 |
Table 3 Relationship Between PRDX1 Expression and Clinicopathological Features |
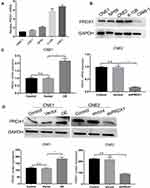
PRDX1 Expression in Human NPC Cell Lines
Next we chose four NPC cell lines (CNE1, CNE2, 6–10B, and C666-1) and one nasopharyngeal normal cell line (NP69) to examine PRDX1 expression. PRDX1 expression was higher in CNE1 and C666-1 than in NP69, and PRDX1 expression in CEN2 was similar to the expression in NP69 (Figure 2A and B). Therefore, we chose to upregulate PRDX1 expression in CNE1 and silence PRDX1 expression in CNE2 to explore biological function of PRDX1 in NPC cells. We confirmed that PRDX1 mRNA and protein levels were significantly higher in CNE1 cells infected with lentivirus harboring PRDX1 cDNA, but were significantly lower in CNE2 cells infected with lentivirus harboring PRDX1 shRNA (Figure 2C and D).
PRDX1 Inhibits NPC Cell Proliferation, Migration, and Invasion
CCK-8 assay showed that overexpression of PRDX1 suppressed CNE1 cell proliferation (Figure 3A), while knockdown of PRDX1 enhanced CNE2 cell proliferation (Figure 3B). Wound healing assay demonstrated that overexpression of PRDX1 suppressed CNE1 cell migration (Figure 3C), while knockdown of PRDX1 enhanced CNE2 cell migration (Figure 3D). Similarly, transwell assay revealed that PRDX1 overexpression decreased CNE1 cell migration, and PRDX1 knockdown enhanced CNE2 cell migration (Figure 3E). In addition, matrigel invasion assay showed that PRDX1 overexpression markedly suppressed CNE1 cell invasion, while PRDX1 silencing enhanced CNE2 cell invasion (Figure 3F). These results suggest that PRDX1 inhibits NPC cell growth and invasion in vitro.
PRDX1 Inhibits EMT of NPC Cells
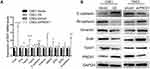
To explore how PRDX1 regulates NPC cell invasion, we detected the expression of EMT related markers which play an mportant role in tumor metastasis.17,18 Real-time PCR showed that PRDX1 overexpression markedly increased the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin, Snail, Slug, Twist1, Twist2, Zeb1, Zeb2, and MMP9 in CNE1 cells, while knockdown of PRDX1 showed opposite results in CNE2 cells (Figure 4A). Notably, the changes of E-cadherin, N-cadherin, Snail, Twist1, and Vimentin expression were confirmed by Western blot analysis (Figure 4B). Taken together, these data indicate that PRDX1 inhibits EMT of NPC cells.
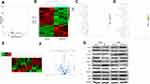
Transcriptome of NPC Cells with PRDX1 Knockdown
To elucidate molecular mechanisms underlying PRDX1 function in NPC, we performed transcriptome sequencing analysis in a CNE2-shPRDX1 cell. We found 208 genes differentially expressed in PRDX1 knockdown cells compared to control cells, including 85 upregulated genes and 123 downregulated genes. The volcano map and Heat map analysis further exhibited the change of these genes (Figure 5A and B). GO analysis showed that PRDX1 participated in a multicellular organismal process (GO: 0032501), epithelium development (GO: 0060429), and epithelium cell differentiation (GO: 0030855) (Figure 5C). The KEGG signaling pathway demonstrated the top 20 pathways in enrichment (Figure 5D). We chose the KEGG signaling pathway of small cell lung cancer among these signaling pathways, and found that the PI3K/AKT signaling pathway was enriched after PRDX1 silencing. The heat map of this signaling pathway showed different gene expression between shPRDX1 and control cells (Figure 5E). Furthermore, the volcano map shows the top four genes are significantly related to the PI3K/AKT pathway in CNE2-shPRDX1 cells (Figure 5F).
Furthermore, total protein levels of PI3K, AKT, and p65 were similar in CNE1-OE cells compared to CNE1-Vector cells, but the phosphorylation of PI3K (P-PI3K), AKT (P-AKT), p65 (P-p65), and TRAF1 significantly decreased in PRDX1 overexpressing cells (Figure 5G). Consistently, PRDX1 knockdown led to increased phosphorylation of PI3K (P-PI3K), AKT (P-AKT), p65 (P-p65), and TRAF1 in CNE2 cells (Figure 5G). Taken together, these results suggest that PRDX1 regulates the PI3K/AKT/TRAF1 pathway in NPC cells.
PRDX1 Inhibits NPC Growth in vivo
Finally, we established a tumor-bearing nude mice model loaded with CNE1-OE cells and CNE2-shPRDX1 cells to monitor the effect of PRDX1 on cancer growth in vivo. The results showed that PRDX1 overexpression reduced tumor volume, while PRDX1 knockdown increased tumor volume (Figure 6A–D). After 4 weeks, the tumors from the four mice were collected, and PRDX1 expression level was confirmed at protein level (Figure 6E and F). Collectively, these results indicate that PRDX1 could inhibit NPC growth in vivo.
Discussion
Despite molecular targeting treatment the prognosis of NPC patients is still poor. Therefore, the identification of biomarkers and understanding the mechanisms of NPC may provide a new strategy for NPC diagnosis and therapy. PRDX1 has been reported to be implicated in cancer development.19 In this study, we performed immunohistochemical analysis to show that PRDX1 expression was low in NPC samples, and PRDX1 expression level was negatively associated with local recurrence and distant metastasis in NPC tissues.
In papillary thyroid cancer, PRDX1 was significantly reduced and acted as a tumor suppressor via BRAF V600E-dependent mechanism.20 BRAF V600E could inhibit PRDX1 expression in papillary thyroid cancer cells.20 We speculated that PRDX1 may inhibit NCP metastasis by suppressing EMT, a crucial process in cancer metastasis.21 Our results showed that silencing PRDX1 expression could promote NPC cell proliferation, migration, and invasion, while overexpression of PRDX1 could inhibit NPC cell proliferation, migration, and invasion. Based on the analysis of the changes of EMT-related factors, we confirmed that PRDX1 could suppress EMT of NPC cells.
PI3K could regulate NF-kB binding to DNA sequence via the activation of AKT, which in turn mediates EMT.22 By transcriptome sequencing analysis, we showed that PRDX1 regulated downstream targets of PI3K/AKT signaling. In addition, silencing PRDX1 could increase the phosphorylation of PI3K, AKT, and p65. These data suggest that PRDX1 may regulate EMT process in NPC cells through regulating PI3K/AKT/NF-κB signaling.
TRAF1 plays a diverse role in regulating gene expression and cellular activities in many tumors.23 In non-small cell lung cancer, the expression of anti-apoptotic protein TRAF1 was upregulated, and the PI3K/AKT/NF-κB pathway was activated.24 In this study, TRAF1 was the most important downstream of PI3K/AKT signaling in NPC cells based on the analysis of the KEGG signaling pathway. Moreover, silencing PRDX1 could increase TRAF1 protein expression in NPC cells. Taken together, we speculate that PRDX1 could regulate TRAF1 downstream of PI3K/AKT signaling pathway. Further studies are needed to reveal the mechanism by which PRDX1 is downregulated in NPC.
In conclusion, our findings reveal PRDX1 as a tumor suppressor in human NPC. The expression of PRDX1 was negatively associated with recurrence and metastasis of NPC after treatment. PRDX1 inhibits NPC cell proliferation, migration, and invasion via suppressing the PI3K/AKT/TRAF1 signaling pathway. These results suggest that PRDX1 may be a new prognostic indicator and therapeutic target in NPC.
Author Contributions
Conception and design: Hongmei Xiao, Jihong Feng, Yu Jiang. Provision of materials: Hongmei Xiao, Taoyu Yang, Lingli Yan, Jihong Feng, Boyan Huang. Collection and assembly of data: Hongmei Xiao, Taoyu Yang, Lingli Yan, Jihong Feng, Boyan Huang. Data analysis and interpretation: Hongmei Xiao, Taoyu Yang, Lingli Yan. Manuscript writing: All authors. Final approval of manuscript: All authors. Accountable for all aspects of the work: All authors.
Funding
This study was supported by grants from Guizhou Science and Technology Department (No. QKH-LH-2015-7505, QKH-RC-2019-5612).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Fan C, Tang Y, Wang J, et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9:2852–2864. doi:10.7150/jca.25460
2. Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi:10.1016/S0140-6736(19)30956-0
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi:10.3322/caac.21551
4. Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019;39:22. doi:10.1186/s40880-019-0368-6
5. Zhao CX, Zhu W, Ba ZQ, et al. The regulatory network of nasopharyngeal carcinoma metastasis with a focus on EBV, lncRNAs and miRNAs. Am J Cancer Res. 2018;8:2185–2209.
6. Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol Med. 2005;11:571–578. doi:10.1016/j.molmed.2005.10.006
7. Wang X, He S, Sun JM, Delcuve GP, Davie JR. Selective association of peroxiredoxin 1 with genomic DNA and COX-2 upstream promoter elements in estrogen receptor negative breast cancer cells. Mol Biol Cell. 2010;21:2987–2995. doi:10.1091/mbc.e10-02-0160
8. Ren P, Ye H, Dai L, et al. Peroxiredoxin 1 is a tumor-associated antigen in esophageal squamous cell carcinoma. Oncol Rep. 2013;30:2297–2303. doi:10.3892/or.2013.2714
9. Kim YJ, Lee WS, Ip C, Chae HZ, Park EM, Park YM. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66:7136–7142. doi:10.1158/0008-5472.CAN-05-4446
10. Chhipa RR, Lee KS, Onate S, Wu Y, Ip C. Prx1 enhances androgen receptor function in prostate cancer cells by increasing receptor affinity to dihydrotestosterone. Mol Cancer Res. 2009;7:1543–1552. doi:10.1158/1541-7786.MCR-08-0546
11. Cai AL, Zeng W, Cai WL, et al. Peroxiredoxin-1 promotes cell proliferation and metastasis through enhancing AKT/mTOR in human osteosarcoma cells. Oncotarget. 2018;9:8290–8302.
12. Ha B, Kim EK, Kim JH, et al. Human peroxiredoxin 1 modulates TGF-β1-induced epithelial-mesenchymal transition through its peroxidase activity. Biochem Biophys Res Commun. 2012;421:33–37. doi:10.1016/j.bbrc.2012.03.103
13. Guo QJ, Mills JN, Bandurraga SG, et al. MicroRNA-510 promotes cell and tumor growth by targeting peroxiredoxin1 in breast cancer. Breast Cancer Res. 2013;15:R70. doi:10.1186/bcr3464
14. Riddell JR, Maier P, Sass SN, Moser MT, Foster BA, Gollnick SO. Peroxiredoxin 1 stimulates endothelial cell expression of VEGF via TLR4 dependent activation of HIF-1α. PLoS One. 2012;7:e50394. doi:10.1371/journal.pone.0050394
15. Lu J, Huang H, Zeng Q, et al. Hippocampal neuron loss and astrogliosis in medial temporal lobe epileptic patients with mental disorders. J Integr Neurosci. 2019;18(2):127–132.
16. Yang C, Tian Y. SPAG9 promotes prostate cancer growth and metastasis. Biocell. 2019;43(3):207–214. doi:10.32604/biocell.2019.07258
17. Terry S, Savagner P, Ortiz-Cuaran S, et al. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11:824–846. doi:10.1002/1878-0261.12093
18. Wu T, Bao T, Liang D, Wang L. Epithelial-mesenchymal transition contributes to malignant phenotypes of circulating tumor cells derived from gastric cancer. Biocell. 2019;43(4):293–298. doi:10.32604/biocell.2019.07841
19. Ding C, Fan X, Wu G. Peroxiredoxin 1 - an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21(1):193–202. doi:10.1111/jcmm.12955
20. Nicolussi A, D’Inzeo S, Mincione G, et al. PRDX1 and PRDX6 are repressed in papillary thyroid carcinomas via BRAF V600E-dependent and -independent mechanisms. Int J Oncol. 2014;44(2):548–556. doi:10.3892/ijo.2013.2208
21. Guo Q, Huang Y, Xu J, et al. Long non-coding RNA BANCR contributes to cervical adenocarcinoma migration by affecting epithelial-mesenchymal transition. Eur J Gynaecol Oncol. 2019;40(3):408–412.
22. Shih MC, Chen JY, Wu YC, et al. TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene. 2012;31(19):2389–2400. doi:10.1038/onc.2011.419
23. Edilova MI, Abdul-Sater AA, Watts TH. TRAF1 signaling in human health and disease. Front Immunol. 2018;9:2969. doi:10.3389/fimmu.2018.02969
24. Song L, Xiong H, Li J, et al. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-kB pathway in human non–small cell lung cancer. Clin Cancer Res. 2011;17(7):1839–1849. doi:10.1158/1078-0432.CCR-10-0720
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.