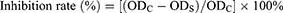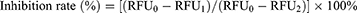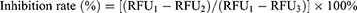Back to Journals » Journal of Inflammation Research » Volume 15
Potential Multifunctional Bioactive Compounds from Dysosma versipellis Explored by Bioaffinity Ultrafiltration-HPLC/MS with Topo I, Topo II, COX-2 and ACE2
Authors Feng H, Chen G, Zhang Y, Guo M
Received 22 April 2022
Accepted for publication 22 July 2022
Published 15 August 2022 Volume 2022:15 Pages 4677—4692
DOI https://doi.org/10.2147/JIR.S371830
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Huixia Feng,1– 4 Guilin Chen,1– 4 Yongli Zhang,1– 4 Mingquan Guo1– 4
1Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, 430074, People’s Republic of China; 2University of Chinese Academy of Sciences, Beijing, 100049, People’s Republic of China; 3Sino-Africa Joint Research Center, Chinese Academy of Sciences, Wuhan, 430074, People’s Republic of China; 4Innovation Academy for Drug Discovery and Development, Chinese Academy of Sciences, Shanghai, 201203, People’s Republic of China
Correspondence: Mingquan Guo, Tel +86-02787700850, Email [email protected]
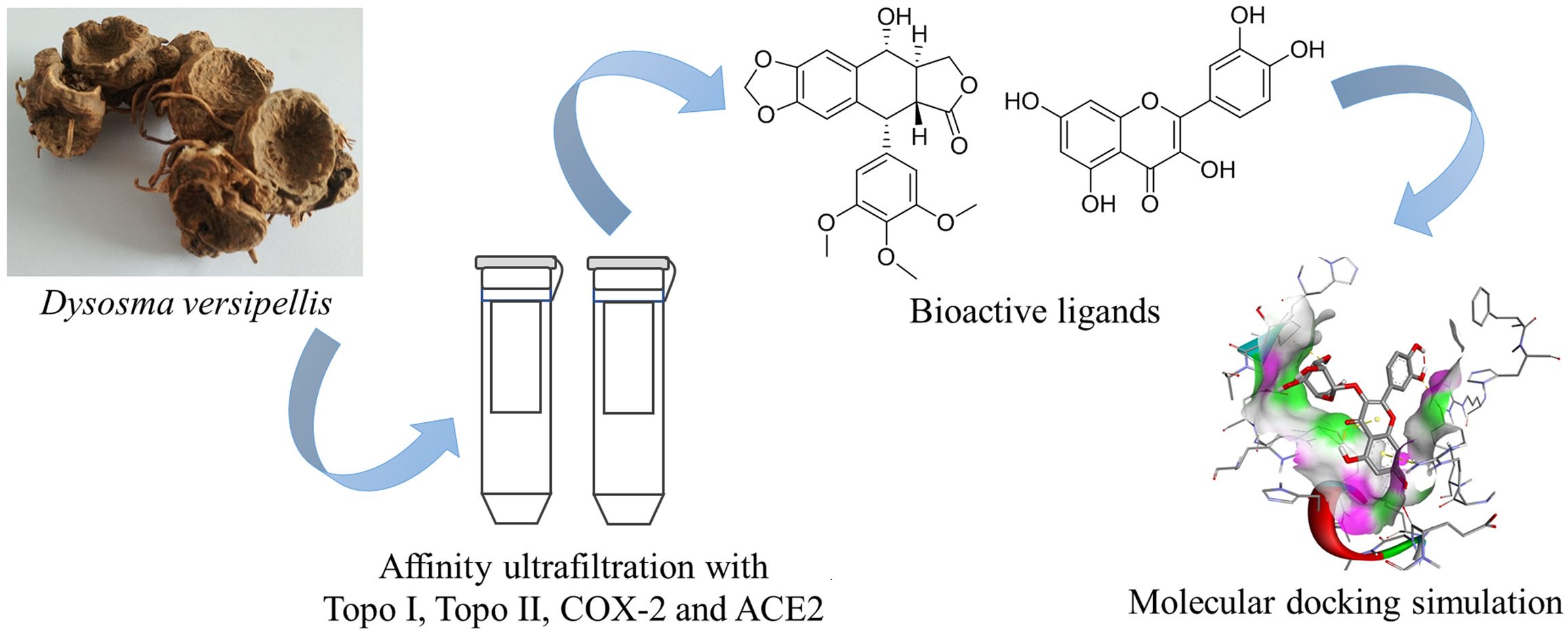
Background: Dysosma versipellis (D. versipellis) has been traditionally used as a folk medicine for ages. However, the specific phytochemicals responsible for their correlated anti-inflammatory, anti-proliferative and antiviral activities remain unknown.
Purpose: This study aimed to explore the specific active components in D. versipellis responsible for its potential anti-inflammatory, anti-proliferative, and antiviral effects, and further elucidate the corresponding mechanisms of action.
Methods: Bioaffinity ultrafiltration coupled to liquid chromatography–mass spectrometry (UF-LC/MS) was firstly hired to fast screen for the anti-inflammatory, anti-proliferative and antiviral compounds from rhizomes of D. versipellis, and then further validation was conducted using in vitro inhibition assays and molecular docking.
Results: A total of 12, 12, 9 and 12 phytochemicals with considerable affinities to Topo I, Topo II, COX-2 and ACE2 were fished out, respectively. The anti-proliferative assay in vitro indicated that podophyllotoxin and quercetin exhibited comparably strong inhibitory rates on A549 and HT-29 cells compared with 5-FU and etoposide. Meanwhile, kaempferol displayed prominent dose-dependent inhibition against COX-2 with IC50 value at 0.36 ± 0.02 μM lower than indomethacin at 0.73 ± 0.07 μM. Furthermore, quercetin exerted stronger inhibitory effect against ACE2 with IC50 value at 104.79 ± 8.26 μM comparable to quercetin 3-O-glucoside at 135.25 ± 6.54 μM.
Conclusion: We firstly showcased an experimental investigation on the correlations between bioactive phytochemicals of D. versipellis and their multiple drug targets reflecting its potential pharmacological activities, and further constructed a multi-target and multi-component network to decipher its empirical traditional applications. It could not only offer a reliable and valuable experimental basis to better comprehend the curative effects of D. versipellis but also provide more new insights and strategies for other traditional medicinal plants.
Keywords: ultrafiltration, Dysosma versipellis, multiple drug targets, anti-inflammatory, anti-proliferative, antiviral
Graphical Abstract:
Introduction
Dysosma versipellis (Hance) M. Cheng ex Ying belongs to Dysosma genus, Berberidaceae Family. It has a long history of application in the folk with extremely high medicinal value for the treatment of traumatic injuries, rheumatic pain and snake bites.1,2 This medical plant was first recorded in “Shen-Nong-Ben-Cao-Jing” and was mainly distributed in Guizhou, Sichuan, Yunnan and Hubei provinces of China.3 The major phytochemicals in D. versipellis were aryltetralin-type lignans, like podophyllotoxin, with extensive physiological activities including anti-proliferative, anti-inflammatory, antiviral, and antibacterial effects.3,4 At present, the majority of research on D. versipellis mainly focuses on the extraction and separation of chemical components, content determination, and structural modification of compounds.5,6 However, some challenges have not been settled yet, such as which chemical components exert pharmacological effects in D. versipellis, how they act, and how their mechanisms work. It has become a bottleneck to efficiently single out bioactive components responsible for their highly specific activities from D. versipellis, and further decipher its underlying mechanism of action.
Most of the new drug research in the past few decades has concentrated on finding or designing highly selective drug molecules that act on a single target. However, for some complex multi-cause diseases like central nervous system, cardiovascular and cerebrovascular diseases, single-target drugs are difficult to interfere with the complex regulatory network of the diseases, and achieve the desired therapeutic effects, even causing unforeseen side effects.7 In this regard, multi-target drugs provide brand-new thinking for drug discovery. Compared with single-target drugs, multi-target drugs can act on multiple targets that are inherently related to corresponding diseases, and regulate multiple signaling pathways of the disease.8 Due to the synergistic actions between multiple effector molecules, the total effects are greater than the sum of the individual effects, which could thus increase the curative effect, reduce adverse reactions and improve drug resistance.9 Generally speaking, the interaction between small drug molecules and target biomacromolecules like enzymes in vivo is one of the critical steps for most of the drugs to exert their pharmacological effects.10,11 Given the diversified pharmacological activities of D. versipellis in many aspects, DNA topoisomerase I (Topo I), DNA topoisomerase II (Topo II), cyclooxygenase-2 (COX-2), and angiotensin-converting enzyme 2 (ACE2) closely correlated to the empirical applications of D. versipellis were selected in the present study as the critical drug targets responsible for its anti-proliferative, anti-inflammatory and antiviral activities, respectively. Among these targets, DNA topoisomerases (Topo), a kind of ribozyme, are classified into types I and II based on their catalytic function. Topo I clears DNA single-strand, whereas Topo II breaks double-strands.12 Plentiful anti-cancer drugs, including campodeids of hydroxycamptothecin and irinotecan, podophyllin derivatives of etoposide and teniposide, exert their therapeutic effects through Topo I and Topo II by interfering with DNA replication, recombination and gene expression, respectively.13 Therefore, it has become a new hot spot to design various Topo inhibitors, and transform them into clinically relevant anti-neoplastic agents.14 With regard to COX-2, it is an essential rate-limiting enzyme in the conversion of arachidonic acid (AA) to prostaglandins (PGs). Moreover, it catalyzes the generation of PGs that participates in various physiological and pathophysiological processes of the body, such as inflammation, apoptotic resistance, angiogenesis, proliferation, invasion, and metastasis of cancer cells.15,16 Hence, it serves as an essential target for the development of novel anti-inflammatory and auxiliary anti-proliferative drugs.17 Concerning ACE2, it has recently been considered as the SARS-CoV-2 receptor with a link between inflammation, immunity and cardiovascular diseases.18–20 Thereinto, ACE2 is down-regulated after SARS-CoV-2 virus infection, and numerous inflammatory signals will trigger a cytokine storm, thereby aggravating inflammation.21,22 Meanwhile, ACE2 counterbalances multiple functions as a powerful negative regulator of the renin–angiotensin system (RAS). In addition, it also relates closely to the expression of amino acid transporters in the kidneys and guts.22
To a certain extent, the efficacy of the medicine gives rise to distinct and intricate pharmacological mechanisms due to the characteristics of its chemical composition.23 Therefore, it becomes an essential factor to figure out the bioactive constituents, and to further elucidate their potential mechanisms of action in the study of natural products. Bioaffinity ultrafiltration mass spectrometry (UF-LC/MS) approach could quickly screen and identify bioactive components from natural products.24,25 Accordingly, this technology could not only contribute to unravelling the mechanisms of action for bioactive small molecules but also provide novel approaches and ideas for the study of natural products.24 Enlightened by the above observations, the present study aims to comprehensively explore the specific phytochemicals responsible for anti-proliferative, anti-inflammatory and antiviral activities in the traditional applications of D. versipellis employing UF-LC/MS technology combined with four drug targets (Topo I, Topo II, COX-2, and ACE2). To the best of our knowledge, our present work for the first time introduced the UF-LC/MS strategy with four drug targets for the high-throughput screening and rapid characterization of their respective ligands from D. versipellis. Herein, we further constructed a ligand–target interacting network between multi-components and their respective multi-targets based on those experimental findings, which could be very conducive to unveiling its underlying mechanisms of the empirical traditional applications of D. versipellis.
Materials and Methods
Plant Material
Fresh rhizomes of D. versipellis were collected in 2013 from the Shennongjia forest area in Hubei, China. The authentication of this specimen was generously helped by Professor Guangwan Hu, a plant taxonomist of the Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences. The rhizome of D. versipellis was crushed into powder and then stored in sealed polyethylene bags at 4 ℃ until use. The voucher specimen (No. 20190701) has been deposited.
Chemicals and Reagents
DNA Topo I and Topo II were purchased from New England Biolabs (Ipswich, MA, USA). Recombinant human COX-2 and ACE2 were bought from Sigma-Aldrich (St Louis, MO, USA) and Novoprotein (Shanghai, China), respectively. Indomethacin, 5-fluorouracil (5-FU), kaempferol and podophyllotoxin were provided by Shanghai Aladdin Bio-Chem Technology Co., Ltd. (Shanghai, China). Etoposide was obtained from Shanghai Yuanye Bio-technology Co., Ltd. (Shanghai, China). The reference standards of kaempferol 3-O-glucoside, quercetin and quercetin 3-O-glucoside were obtained from Shanghai Tauto Biotech Co., Ltd. (Shanghai, China). The centrifugal ultrafiltration filters of 30 kDa (YM-30) were provided by Millipore Co., Ltd. (Bedford, MA, USA). Sulforhodamine B (SRB) cell proliferation and cytotoxicity detection kits, COX-2 inhibitor screening assay kits and ACE2 inhibitor screening assay kits were obtained by Shanghai Beyotime Biotechnology Co., Ltd. (Shanghai, China). The Millipore membranes (0.22 µm) were bought from Tianjin Jinteng Experiment Equipment Co., Ltd. (Tianjin, China). The HPLC-grade solvents of formic acid (FA) and acetonitrile (ACN) were obtained from TEDIA Company Inc. (Fairfield, OH, USA). The ultra-pure water for all solutions, dilutions and HPLC-ESI-MS analysis was generated by EPED water system from Yeap Esselte Tech. Co., Ltd. (Nanjing, China). All other chemicals and solvents for analysis, unless specified, were bought from Shanghai Chemical Reagent Corp. (Shanghai, China).
Preparation of Sample Crude Extract
With respect to the sample preparation, an aliquot of 200 g raw powders of D. versipellis was extracted ultrasonically 3 times with 90% ethanol at room temperature for 30 min. The extract was then concentrated utilizing a reduced pressure evaporator, and finally freeze-dried to obtain the crude extract of D. versipellis.
Affinity Ultrafiltration Screening
The present affinity ultrafiltration screening procedures were implemented according to the previous literature with slight modifications.26,27 Briefly, the D. versipellis extract was dissolved in phosphate buffer saline (PBS, pH 7.4) to prepare the sample solution for ultrafiltration screening. 100 µL of the sample solution (at a final concentration of 8 mg/mL) was mixed with 10 µL of Topo I (5 U), Topo II (2 U), COX-2 (5 U) or ACE2 (0.5 µg), and incubated at 37 ℃ for 40 min. The mixtures were filtered through a 30 KD cut-off ultrafiltration membrane at 10,000 rpm for 10 min. Afterward, unbound components were removed by eluting the filtrates 3 to 4 times with 200 µL of PBS (pH 7.4) by centrifugation at 10,000 rpm for 10 min. Furthermore, 200 µL of 90% methanol was added to the mixed filtrates to release bound ligands. After incubating at room temperature for 10 min, the filtrates were centrifuged at 10,000 rpm for 10 min by repeating 3 to 4 times. Finally, the eluted filtrates were re-dissolved in 50 µL of methanol after lyophilizing and later subjected to HPLC-ESI-MS/MS analysis. In the meantime, the inactivated enzyme solution after being placed in boiling water for 10 min was considered as the control group.
HPLC-MS Analysis
The HPLC analysis was performed on a Thermo Access 600 HPLC system (Thermo Fisher Scientific, San Jose, CA, USA). Briefly, a Waters Symmetry RP-C18 column (4.6 mm × 250 mm, 5 µm) with a guard column was served for the chromatographic separations. The mobile phases consisted of 0.1% FA – H2O (A) and ACN (B). The separation was conducted with the optimized elution conditions as follows: 0 – 40 min, 5 – 95% B. The samples (10 µL) were injected into the system with a flow rate of 0.8 mL/min at 30 ℃, and the eluent was monitored at a wavelength of 292 nm.
As for the ESI-MS/MS analysis, the mass spectrometer with electrospray ionization (ESI) was operated to obtain diverse fragment ions in the positive ion mode with the spray voltage of 3.0 kV and the cone voltage of 40.0 V; the vaporizer temperature and capillary temperature were set at 350 ℃ and 250 ℃, respectively; the sheath gas pressure and aux gas pressure were recognized as 40 psi and 10 psi, respectively; the scan range was m/z 150 Da to 1000 Da; the collision energy altered from 30 eV to 45 eV for the MS/MS analysis. All the analytical data was obtained from the professional software of Thermo Xcalibur ChemStation (Thermo Fisher Scientific).
Anti-Proliferative Assay
The anti-proliferative assay in vitro on non-small lung cell cancer (A549) and colon cancer (HT-29) cell lines was performed by applying a SRB kit according to the manufacturer’s instructions with minor modification.28 These cell lines were purchased commercially. During the procedures of anti-proliferative assay, these two cell lines were seeded into the 96-well plates with a density of 1.0 × 104 per well maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin, and incubated in a humidified atmosphere comprising 5% CO2 at 37 ℃ for 24 h. Then, the cells were covered with 100 µM of the tested samples dissolved in dimethyl sulfoxide (DMSO). 5-FU as well as etoposide were considered as the positive controls, and less than 0.1% DMSO was regarded as the blank control. In the end, the optical density (OD) value of each well was detected at 540 nm with a multifunctional microplate reader (Tecan Infinite M200 PRO, TECAN, Männedorf, Switzerland). The inhibition rate (%) was computed in accordance with Eq. (1):
where ODC and ODS represent the absorbance values of the blank control and the tested samples or positive controls, respectively. All the sample solutions were measured in triplicate.
COX-2 Inhibitory Assay
The COX-2 inhibitory assay in vitro was carried out to measure the anti-inflammatory activity of representative compounds with a COX-2 inhibitor screening kit referring to the previous approaches with slight modifications.29 Before use, the prepared COX-2 cofactor solution, COX-2 solution, COX-2 substrate solution and COX-2 probe solution were diluted 10 times with COX-2 assay buffer. Briefly, 150 µL of tri (hydroxymethyl) aminomethane hydrochloride (Tris-HCl, pH 7.9), 10 µL of COX-2 cofactor solution and 10 µL of COX-2 solution were firstly mixed in a 96-well blackboard, and 10 µL of tested sample solution with the concentrations of 0.625–20 µM was added to each mixture, which was then incubated at 37 ℃ for 10 min. Afterward, 10 µL of COX-2 probe solution and 10 µL of COX-2 substrate solution were quickly appended to the mixtures, and finally fluorescence detection was carried out after incubation in the dark for 5 min at 37 ℃. The emission wavelength was determined at 590 nm, and the excitation wavelength of the reaction mixtures was monitored at 560 nm. Indomethacin was considered as the positive control. The inhibition rate (%) of each sample was calculated, as shown in Eq. (2):
where RFU0 is the relative fluorescence unit of the 100% enzyme activity control group, RFU1 is the relative fluorescence unit of the tested sample, and RFU2 is the relative fluorescence unit of the blank control group, respectively. Each sample solution was measured in three parallels. And the final experimental results were conveyed in the form of mean ± standard deviation (SD). The half-maximal inhibitory concentration (IC50) value was computed by constructing the inhibition rates against the tested sample.
ACE2 Inhibitory Assay
The ACE2 inhibitory assay in vitro was implemented to detect the antiviral activity of several components screened out with an ACE2 inhibitor screening kit based on the manufacturer’s instructions and the previous studies.29 Before use, the prepared ACE2 solution and substrate were diluted 5 times with ACE2 assay buffer. Briefly, 92 µL of PBS (pH 6.8), 1 µL of ACE2 solution and 5 µL of tested sample solution or the positive drug were firstly mixed in a 96-well blackboard. Then, it was incubated at 37 ℃ for 15 min. Afterward, 2 µL of ACE2 substrate solution was quickly added to the mixtures under low temperatures, and finally the fluorescence detection was performed after incubation in the dark for at least 2 h at 37 ℃. The emission wavelength was measured at 325 nm, and the excitation wavelength was detected at 393 nm. The inhibition rate (%) of each sample was computed applying Eq. (3) as follows:
In the above formula, RFU1, RFU2 and RFU3 represent the relative fluorescence unit of the 100% enzyme activity control group, the relative fluorescence unit of the tested sample, and the relative fluorescence unit of the blank control group, respectively. All the sample solutions were detected in triplicate, and the results were expressed by mean ± SD.
Molecular Docking Assay
The molecular docking assay between potential bioactive ligands and Topo I, Topo II, COX-2 or ACE2 was carried out on the basis of the published literature previously.29 Briefly, the three-dimensional structures of Topo I (PDB ID: 1T8I), Topo II (PDB ID: 3QX3), COX-2 (PDB ID: 1CX2) and ACE2 (PDB ID: 1R42) and potential bioactive ligands with the lowest energies were downloaded and established from the PDB databases and ChemBio3D Ultra 14.0, respectively. Both files were saved in PDB format. Next, the receptor and ligands were subjected to structural processing with AutodockTools software, including removing the water molecular, adding the hydrogen atoms, charging treatment, and minimizing in MMFF94 × force field. Both files were converted to pdbqt files. Afterwards, the Grid dialog box was set with the centroid coordinate as follows: Topo I (X_21.474; Y_-2.226; Z_27.863), Topo II (X_33.026; Y_95.765; Z_51.567), COX-2 (X_24.263; Y_21.528; Z_16.497) or ACE2 (X_52.874; Y_68.399; Z_33.501), and the three-dimensional size was adjusted as 60 × 60 × 60 through the knob. After that, the configuration file was saved and exported. Subsequently, molecular docking processes between the receptor and the ligand were performed with 2.5 × 106 energy evaluations as well as 50 independent runs of the Genetic Algorithm. Finally, the best orientation was confirmed by linking potential bioactive ligands to target proteins with the lowest binding energy. All the docking results were acquired and visualized by adopting the AutoDockTools 1.5.6 and Discovery Studio 4.5 Client.
Statistical Analysis
All the assays were performed in triplicate. All statistical data analysis in the present study was conducted with the SPSS 16.0 Software (SPSS Inc., Chicago, IL, USA), the Origin 2019 Software (OriginLab Corp., Northampton, MA, USA), the Chemoffice 14.0 Software (CambridgeSoft Corp., Cambridge, MA, USA) and the GraphPad Prism 8 Software (GraphPad Software Corp., San Diego, CA, USA). Results were expressed as mean ± SD, and the differences were considered statistically significant at P < 0.05.
Results and Discussion
Identification of Topo I, Topo II, COX-2 and ACE2 Ligands
As displayed in Figure 1, after the affinity ultrafiltration screening, a total of 12, 12, 9 and 12 phytochemicals from the crude extract of D. versipellis were found to exert different binding affinities to Topo I, Topo II, COX-2 and ACE2, respectively. Those components were simultaneously identified by HPLC-ESI-MS/MS analysis.30 The MS/MS data were detected in the positive ion mode, and the retention times (Rt), quasi-molecular ([M+H]+), and their characteristic fragment ions are summarized in Table 1. Based on the corresponding MS/MS data and fragmentation pathways of several representative compounds reported in the previous literature as well as the spectra of the corresponding standards, 12 relevant bioactive components coupled with Topo I, Topo II, COX-2 and ACE2 from the crude extract of D. versipellis were characterized, and their chemical structures are displayed in Figure 2.
 |
Table 1 Identification, EFs, and MS/MS Data of the Bioactive Components Bound to Topo I, Topo II, COX-2 and ACE2 from the Crude Extract of D. versipellis |
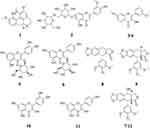 |
Figure 2 Chemical structures of 12 compounds identified from D. versipellis based on the MS/MS fragment information or the corresponding standards. |
Among these bioactive compounds selected, peak 1 presented [M+H]+ ion at m/z 342 and was further suffered from the characteristic losses to obtain a series of fragment ions, such as the [M+H-CH3-OCH3]+ at m/z 296, the [M+H-(CH3)2NH-H2O]+ at m/z 279, the [M+H-(CH3)2-OCH3]+ at m/z 264 and the [M+H-(CH3)2-OCH3-CO]+ at m/z 236. Based on the comparison with the MS/MS data of the previous study, peak 1 was identified as isocorydine (calculated for C20H23NO4, 341 Da).31 Peak 2 exhibited the [M+H]+ ion at m/z 625, and the fragment ions at m/z 463 indicated the loss of a hexose moiety [M+H-162]+. By comparing its MS/MS spectra with the database, peak 2 was tentatively identified as chrysoeriol glucosylglucoside (calculated for C20H23NO4, 341 Da). Peaks 3 and 4 showed the same [M+H]+ ion at m/z 299 and MS/MS spectra at m/z 284, m/z267, m/z239, m/z211, m/z181 and m/z153, implying that they are the structural isomers. Fragment ions at m/z 284, m/z267, m/z239, m/z211, m/z181 and m/z153 were probably produced by a series of neutral losses from the parent ion corresponding to [M+H-CH3]+, [M+H-CH3-OH]+, [M+H-CH3-OH-CO]+, [M+H-CH3-OH-(CO)2]+, [M+H-CH3-OH-(CO)2-CH2O]+ and [M+H-CH3-OH-(CO)3-CH2O]+, respectively. According to the previous literature, peaks 3 and 4 were presumed to be 3’,7-dimethoxy-3-hydroxyflavone isomers.32
As for peak 5, the [M+H]+ ion at m/z 465 was obtained. Typically, the aglycon ion [M+H-162]+ at m/z 303 was formed by the loss of a hexose moiety. Peak 5 was thus confirmed as quercetin 3-O-glucoside (isoquercitrin, calculated for C21H20O12, 464 Da) by comparing the Rt and the MS/MS spectra with the corresponding standard. Considering peak 6, the [M+H]+ ion at m/z 449 was produced, and the aglycone ion at m/z 287 indicated the loss of a hexose moiety, which was speculated to be kaempferol monoglycoside. By comparison with the reference standard, peak 6 was regarded as kaempferol 3-O-glucoside (astragalin, calculated for C21H20O11, 448 Da). Meanwhile, peaks 10 ([M+H]+ at m/z 303) and 11 ([M+H]+ at m/z 287) were characterized as quercetin (calculated for C15H10O7, 302 Da) and kaempferol (calculated for C15H10O6, 286 Da) by comparing their MS/MS data with the corresponding standards, respectively.
Peaks 7 and 12 possessed the same deprotonated molecular ion at m/z 415 and other characteristic fragment ions, such as the ions at m/z 397, m/z 313 and m/z 282 produced by the neutral loss of H2O ([M+H-H2O]+), the characteristic retro Diels–Alder (RDA) cleavage ([M+H-H2O-C4H4O2]+) and the loss of the methoxy moiety ([M+H-H2O-C4H4O2-OCH3]+), respectively. Besides, fragment ions at m/z 247 and m/z 229 were probably obtained by the successive neutral losses of the C6H3(OCH3)3 moiety ([M+H-C6H3(OCH3)3]+) and the H2O moiety ([M+H-C6H3(OCH3)3-H2O]+). In comparison with the ESI-MS/MS spectra and the Rt of the corresponding standard, peak 12 was further assigned as podophyllotoxin (calculated for C22H22O8, 414 Da), and peak 7 was considered to be the isomer of peak 12.29 Taking into consideration the [M+H]+ ion at m/z 397 and its MS/MS fragment ions at m/z 313, m/z282, m/z247 and m/z229 similar to peak 12, peak 8 was suggested as β-apopicropodophyllin (calculated for C22H20O7, 396 Da) compared with those ESI-MS/MS spectra and fragmentation pathways.29 Peak 9 exhibited the [M+H]+ ion at m/z 399, and its fragment ions at m/z 231, m/z 203 and m/z 187 were yielded through a sequence of neutral losses of the C6H3(OCH3)3 moiety ([M+H-C6H3(OCH3)3]+), the CO moiety ([M+H-C6H3(OCH3)3-CO]+) and the CO2 moiety ([M+H-C6H3(OCH3)3-CO2]+), respectively. As a result, peak 9 was deduced as deoxypodophyllotoxin (calculated for C22H22O7, 398 Da) in comparison to its MS/MS spectra and fragmentation pathways with the literature reported previously.33
Screening for Topo I, Topo II, COX-2 and ACE2 Ligands from D. versipellis
Numerous researches demonstrate that D. versipellis, a traditional folk medicine, mainly contains lignans, flavonoids and other components, and possesses a broad spectrum of biological activities such as anti-proliferative, anti-inflammatory, antiviral and antibacterial effects.4,34 The interaction of small-molecule drugs with disease-related biomacromolecules in the body is the primary way to exert their pharmacological effects.24 Therefore, the interaction between small molecules of traditional Chinese medicine (TCM) and biomacromolecules could not only contribute to interpreting the mechanism of action of TCM but also efficiently screen the bioactive ingredients for a certain disease, thereby greatly accelerating the development of new drugs from natural products.35 UF-LC/MS technology has become one of the most puissant strategies, which combines the rapid separation of small-molecule ligands from the target macromolecules by affinity ultrafiltration, and the structural identification of potential bioactive components by LC/MS.36 At present, the application of UF-LC/MS for the screening of small-molecule active substances is mainly for a certain target, and there are very few reports on screening methods that act on multi-target components, which still cannot meet the needs of multi-target screening of TCM. Accordingly, UF-LC/MS method with multi-targets was firstly employed in the present study to quickly single out the potential Topo I, Topo II, COX-2 and ACE2 ligands from D. versipellis.
Figure 1 illustrates the UF-LC analysis of the chemical constituents from the crude extract of D. versipellis. In total, 12, 12, 9 and 12 chemical components with specific bindings to four target enzymes were tentatively regarded as the potential Topo I, Topo II, COX-2 and ACE2 ligands, respectively. By comparing the variation of the HPLC peak areas before and after activation of target enzymes in the UF-HPLC chromatograms, the enrichment factor (EF) value of each compound could be applied to evaluate the affinity binding strength between these potential ligands and Topo I, Topo II, COX-2, or ACE2. The calculation of EF value (%) was expressed as Eq. (4):
where the A1, A2 and A0 represent the peak areas from the crude extract of D. versipellis with activated, inactivated and without enzymes, respectively.29
Table 1 enumerates the EF values of the potential ligands from the crude extract of D. versipellis targeting Topo I, Topo II, COX-2 and ACE2, respectively. For Topo I, peak 12 exhibited the highest EF value of 5.39%, followed by the peak 10 of 4.91%, peak 11 of 4.74%, peak 8 of 4.15%, peak 9 of 3.63%, peak 7 of 2.59%, peak 6 of 1.86%, peak 5 of 1.76%, and so on. Concerning Topo II, peak 12 presented the highest EF value of 9.08%, followed by peak 10 of 6.64%, peak 8 of 6.15%, peak 9 of 4.49%, and so forth. With regard to COX-2, peak 11 displayed the highest EF value of 8.71%, followed by peak 12 of 2.45%, peak 8 of 1.79%, peak 9 of 1.13%, peak 7 of 0.94%, etc. As for ACE2, peak 10 showed the highest EF value of 2.98%, followed by peak 12 of 2.86%, peak 8 of 2.83%, peak 9 of 2.07%, peak 7 of 1.93%, peak 6 of 1.49%, and the like. As previously speculated, the EF values of all the peaks were distinct from each other. Those potential ligands with active enzymes demonstrated greater peak areas and intensity than those with inactivated enzymes. Hence, the data imply there existed competitive interactions between those constituents and Topo I, Topo II, COX-2 and ACE2, respectively.
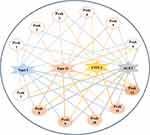
In accordance with the different EF values of constituents screened out and characterized from the crude extract of D. versipellis, the affinities of distinct potential bioactive ingredients to diverse targets can be observed, and used to further figure out enzyme inhibitors with stronger activities. With regard to underlying anti-proliferative activity, peaks 8, 9, 10, 11 and 12 with higher EF values were more retained during the reaction processes due to strong binding affinities with Topo I, and peaks 8, 9, 10 and 12 exhibited higher affinities to Topo II. Obviously, peaks 8, 9, 10 and 12 exerted relatively higher affinities with Topo I and Topo II. These components were tentatively deduced to be the potential bioactive ingredient group, which indicated that there existed synergistic effects among these compounds, conjointly exerting anti-proliferative effects to some extent. Other components with lower EF values were less retained after the reaction courses owing to their lower affinity degrees with Topo I and Topo II. Concerning potential anti-inflammatory activity, peaks 11 and 12 with higher EF values exhibited great binding affinities with COX-2. Meanwhile, peak 11 also showed good binding affinity to Topo I. In regard to the antiviral effect, peak 10 with a higher EF value presented stronger binding affinity with ACE2, which may be served as the dominating antiviral bioactive component. Moreover, the same compounds like peaks 8, 10 and 12 binding to Topo I, Topo II and ACE2 exerted relatively good affinities. These conditions indicated that there existed common bioactive components in D. versipellis for anti-proliferative and antiviral effects. On the one hand, the chemical components with larger EF values could be retained tightly binding to target enzymes during the reaction processes. On the other hand, several potentially bioactive components could target one or multiple target enzymes due to diverse bioactive ligands fished out and characterized. It was found that several active compounds could potentially act on multiple drug targets to exert multiple pharmacological activities, like peaks 8, 10 and 12 for Topo I, Topo II and ACE2, while some constituents displayed a special preference for certain drug targets, like peak 11 for Topo I and COX-2. More strikingly, a multi-target and multi-component interacting network correlated to the empirical applications of D. versipellis was for the first time constructed based on those experimental findings as displayed in Figure 3, which could be very conducive to unveiling the underlying mechanism of the empirical use of D. versipellis, and comprehending in depth the primary curative roles and targets of D. versipellis or other traditional herbal medicines.
In order to determine the accuracy of the screening results, a series of activity verifications in vitro were performed on these selected components from D. versipellis. The anti-proliferative assay in vitro exhibited that peak 10 (quercetin), peak 12 (podophyllotoxin) and peak 11 (kaempferol) with larger EF values manifested relatively strong inhibitory effects (63.24% ± 5.17%, 60.70% ± 0.73% and 60.47% ± 3.99%, respectively) on A549 cells at the concentration of 100 μM, whereas etoposide and 5-FU (positive controls) were 71.13% ± 3.18% and 50.44% ± 1.51%, respectively. Moreover, compared with etoposide and 5-FU (32.24% ± 1.45% and 10.95% ± 1.72%, respectively), quercetin and podophyllotoxin also represented preferable inhibitory activities on HT-29 cells at the concentration of 100 μM, reaching 47.09% ± 3.78% and 46.36% ± 1.47%, respectively. Above all, quercetin was previously provided with favourable anti-proliferative activity on liver cancer (HepG2), gastric carcinoma (SGC-7921) and HT-29 cells with IC50 values at 4.78 ± 0.28 μg/mL, 12.70 ± 0.71 μg/mL and 29.74 ± 0.38 μg/mL, respectively.37 Similarly, kaempferol also exhibited significant inhibition to SGC-7921, HepG2 and HT-29 cells with IC50 values at 13.43 ± 0.55 μg/mL, 20.01 ± 0.97 μg/mL and 25.70 ± 0.82 μg/mL, respectively.38 With regard to the structure-activity relationship of flavonoids, the double bond between C2-C3 on the C ring and the 3’- and 4’- hydroxyl groups on the B ring are essential conditions to ensure the highest anti-proliferative activity of the compounds.37 According to the in vitro tumor cell proliferation inhibition experiments and structure-activity relationship analysis of these two compounds, the results indicated that flavonoids with the double bond between C2-C3 on the C ring and the 3’- and 4’- hydroxyl groups on the B ring possessed higher affinity with Topo I.37 In addition, there existed in corresponding literature reports that podophyllotoxin could inhibit the assembly of microtubules in the mitotic apparatus by preventing the microtubules formed by the polymerization of tubulin, and further destroying the formation of spindle filaments, thereby inhibiting the division and proliferation of tumour cells.39 Studies had shown that new molecular topologies of quercetin complexes could promote DNA cleavage involving single-strand or double-strand breaks which were designed as potential cancer chemotherapeutics.40 Moreover, some studies demonstrated quercetin could strengthen the inhibition of 10-hydroxy camptothecin (HCPT) on the proliferation of human breast cancer cell lines (MCF7), thereby jointly exerting the anti-proliferative mechanism by targeting Topo I to trigger DNA damage and induce cell cycle arrest and apoptosis.41 And molecular docking analysis revealed that quercetin from Ginkgo biloba exhibited a higher affinity potential to interact with Topo II, and it was considered to be the most puissant inducer for DNA damage in HepG2 cells tested by the induction of γ-H2A.X and the Comet assay.42 Furthermore, peak 5 (quercetin 3-O-glucoside) and peak 6 (kaempferol 3-O-glucoside) with relatively low EF values showed lower inhibitory effects on A549 (−4.15% ± 1.69% and −10.66% ± 0.45%) and HT-29 cells (0.57% ± 0.14% and −0.72% ± 0.19%), respectively. As for COX-2, the inhibitory assay in vitro revealed that kaempferol with the highest EF value manifested remarkable dose-dependent inhibition with IC50 value at 0.36 ± 0.02 μM, comparable to indomethacin (positive control) at 0.73 ± 0.07 μM. It was presumably considered to be the potential anti-inflammatory active constituent. Meanwhile, podophyllotoxin with moderate EF value also showed certain inhibition with IC50 value at 10.49 ± 0.61 μM. In addition, kaempferol with a relatively high EF value connected to Topo and COX-2 was speculated that it may act on these two enzymes, so as to exert potential anti-inflammatory and anti-proliferative effects. With respect to ACE2, quercetin with a higher EF value exerted stronger inhibitory effect against ACE2 with IC50 value at 104.79 ± 8.26 μM in contrast to quercetin 3-O-glucoside with relatively lower EF value at 135.25 ± 6.54 μM. The ultrafiltration screening and the in vitro bioactivity inhibitory assay results indicated that this compound was deduced to possess antiviral activity.
Molecular Docking Studies
Research on anti-cancer drugs and anti-inflammatory drugs is a hot topic in the development of new drugs. Meanwhile, after the global outbreak of the coronavirus, the significance of the development of antiviral agents has become prominent. The top priority is to figure out more key targets and develop more anti-proliferative, anti-inflammatory or antiviral drugs. Preliminarily, several phytochemicals possessing potency to target Topo I, Topo II, COX-2 and ACE2 were screened from the crude extract of D. versipellis. Based on their EF values, the screening results in Table 1 revealed that several potent components like podophyllotoxin, quercetin, and kaempferol were scrutinized and tentatively speculated to provide potential anti-proliferative, anti-inflammatory or antiviral effects. Given this situation, in the current study, four phytochemicals (podophyllotoxin, quercetin, kaempferol and quercetin 3-O-glucoside) in D. versipellis were chosen for molecular docking analysis to virtually authenticate the anti-proliferative, anti-inflammatory or antiviral efficacy against the well-recognized drug targets of Topo I, Topo II, COX-2 or ACE2 by simulating the receptor–ligand interactions. The studies on the molecular interaction of receptor–ligand complexes revealed the prevalence of conventional hydrogen bonds (H-bonds), hydrophobic and electrostatic interactions, and non-covalent interactions such as van der Waals forces.43 The intermolecular forces generated by the receptor and the ligand were the inevitable factors that must be considered for the development of successful agents. Several bioactive phytochemicals were listed in detail based on their binding affinities towards Topo I, Topo II, COX-2 and ACE2, as shown in Table 2. Therein, data of molecular docking simulation and the 2D or 3D ligand–target interactions between two phytochemicals and ACE2 are displayed in Figure 4, respectively.
 |
Table 2 The Molecular Docking Results of 4 Phytochemicals Selected from D. versipellis and corresponding positive drugs |
Out of these potential screened ligands, compared with the positive controls 5-FU (−3.7 kcal/mol and 1950.00 μM) and camptothecin (−7.57 kcal/mol and 2.85 μM), podophyllotoxin and quercetin with higher EF values displayed stronger binding affinities to Topo I with its binding energy (BE) of −6.32 kcal/mol and −4.64 kcal/mol as well as the theoretical IC50 value of 23.25 μM and 398.44 μM, respectively. Podophyllotoxin was found to form three conventional H-bonds with amino acid residues Dc112, Lys425 and Met428, and quercetin built five H-bonds with residues Dt10, Leu429, Lys436, Glu356 and Met428. Moreover, these two ligands also exerted stronger binding affinities to Topo II, which their BEs were calculated as −9.45 kcal/mol and −6.99 kcal/mol, and the theoretical IC50 values were 0.12 μM and 7.53 μM, comparable to the positive control etoposide (−7.62 kcal/mol and 2.59 μM). Podophyllotoxin and quercetin were observed to interact with the binding pocket of Topo II to shape H-bonds to enhance binding forces, of which interact with etoposide as well. As regards COX-2, kaempferol and podophyllotoxin depicted higher affinities to it with their BEs of −7.22 kcal/mol and −6.75 kcal/mol as well as the theoretical IC50 values of 5.10 μM and 11.33 μM, which were not much different from the positive control indomethacin (−9.18 kcal/mol and 186.44 nM). Furthermore, kaempferol was discovered to form three stable conventional H-bonds with residues Tyr355, Gln192 and Gly526, and podophyllotoxin constituted one H-bond with residue Arg513 to strengthen the forces. With respect to the antiviral activity, therein, quercetin with the highest EF value topped the list and put forth an inhibitory efficacy against ACE2. The simulation results divulged that its BE and the theoretical IC50 value were −5.17 kcal/mol and 162.36 μM, lower than ACE2 inhibitor MLN-4760 (−4.27 kcal/mol and 738.62 μM), respectively. It was found to exhibit profound interaction with binding pockets to build three conventional H-bonds with residues Ala348, Glu375 and Glu398. In addition, the BE of quercetin 3-O-glucoside with a relatively low EF value was calculated as −4.39 kcal/mol, and its theoretical IC50 value was deduced as 609.37 μM. It underwent four conventional H-bonds with Tyr515, Glu375, Glu398 and Pro346 residues. The docking results indicated that quercetin was equipped with stable interactions between amino acid residues connected to the docking active site of ACE2, and further confirmed the primary roles of this component in D. versipellis for promising antiviral effects. The H-bonds, van der Waals force, hydrophobic and electrostatic effects may facilitate the interaction strengths between ACE2 and those bioactive ligands in the simulation processes. Analogical studies revealed that quercetin possessed good inhibitory potential against ACE2, which its BE was lower than that of the positive control MLN-4760 in the processes of molecular docking.44 There were related literature reports that quercetin and vitamin C co-administration were applied for prophylaxis in high-risk populations and the early treatment of respiratory tract infection (RTI) like COVID-19 patients on account of immunomodulatory and enhanced antiviral effects.45 Quercetin had the characteristics of interfering and blocking virus entry, virus replication and protein assembly with extensive antiviral activities, which combined vitamin C to exert a synergistic antiviral action.45 It was predicted that quercetin could exert the effect on SARS-CoV-2 through interacting with various target proteins such as 3CLpro, PLpro, or S protein based on molecular docking analysis.46 In the meantime, quercetin exhibited remarkable activity against herpes simplex virus type 2 (HSV-2) infection by inhibiting NF-κB activation, markedly lessened the mRNA and protein levels of monocyte chemoattractant protein-1 (MCP-1), interleukin-6 (IL-6), interleukin-8 (IL-8), or intercellular adhesion molecule-1 (ICAM-1) in interleukin-1β (IL-1β)-stimulated human retinal pigment epithelial (ARPE-19) cells, and targeted Ebola virus (EBOV) VP24 anti-type-I Interferon (IFN-I) function to effectively block viral infection as the inhibitor of EBOV VP24 anti-IFN-I function.47–49 In addition, quercetin worked effectively for the intracellular replication and the infectivity of polio-virus type 1 (PV-1), respiratory syncytial virus (RSV), parainfluenza virus type 3 (Pf-3) and herpes simplex virus type 1 (HSV-1).50 Overall, these active compounds screened out may be served as Topo I, Topo II, COX-2 or ACE2 inhibitors and have the potential to develop into anti-proliferative, anti-inflammatory or antiviral drug candidates.
Conclusion
D. versipellis, as a typical Chinese herbal medicine, possessed diversified chemical components and multiple pharmacological activities. Meanwhile, some bioactive phytochemicals in herbal medicine could generate synergistic effects to achieve favourable therapeutic effects in a multi-target manner. Based on the empirical applications of D. versipellis, UF-LC/MS strategy closely combining Topo I, Topo II, COX-2 and ACE2 of interest was successfully developed for the screening and identification of bioactive ligands responsible for anti-proliferative, anti-inflammatory and antiviral activities, which could rapidly filter out the ineffective ones and recognize the targeted bioactive constituents. Next, several bioactive phytochemicals screened out from D. versipellis were further verified by other in vitro bioactivity assays and molecular docking simulation, and the results indicated that the inhibitory effects of some ligands screened were even better than those of positive drugs. It was obvious that this integrative strategy exhibited very promising and considerable potential to rapidly screen and identify multiple bioactive phytochemicals correlated to four respective drug targets selected, which could provide convincing experimental evidences for the empirical applications of D. versipellis. More strikingly, we for the first time inferred and constructed an intricate ligand–target interacting network between multi-components and its corresponding multi-targets, which could be beneficial to unveil the underlying mechanism of D. versipellis. To conclude, we showcased the first systematic and reliable experimental investigation on the correlations between bioactive phytochemicals and their multiple drug targets reflecting multiple pharmacological activities of D. versipellis, which could not only offer a reliable and valuable experimental basis to better comprehend the curative effects of D. versipellis, but also provide some new insights and strategies for other traditional medicinal plants.
Acknowledgments
The authors would also like to thank Prof. Guangwan Hu for his help in identifying the plant material.
Funding
This study was partly supported by the Youth Innovation Promotion Program of the Chinese Academy of Sciences (Grant No. 2020337).
Disclosure
Prof. Mingquan Guo reports a patent 202210029382.5 pending to Mingquan Guo, Huixia Feng, Guilin Chen. The authors declare no other conflicts of interest.
References
1. Liu CX, Zhang CN, He T, et al. Study on potential toxic material base and mechanisms of hepatotoxicity induced by Dysosma versipellis based on toxicological evidence chain (TEC) concept. Ecotoxicol Environ Saf. 2020;190:110073. doi:10.1016/j.ecoenv.2019.110073
2. Man SL, Gao WY, Wei CL, et al. Anticancer drugs from traditional toxic Chinese medicines. Phytother Res. 2012;26(10):1449–1465. doi:10.1002/ptr.4609
3. Tan XM, Zhou YQ, Zhou XL, et al. Diversity and bioactive potential of culturable fungal endophytes of Dysosma versipellis; a rare medicinal plant endemic to China. Sci Rep. 2018;8(1):5929. doi:10.1038/s41598-018-24313-2
4. Jiang RW, Zhou JR, Hon PM, et al. Lignans from Dysosma versipellis with inhibitory effects on prostate cancer cell lines. J Nat Prod. 2007;70(2):283–286. doi:10.1021/np060430o
5. Tang YJ, Zhao W, Li HM. Novel tandem biotransformation process for the biosynthesis of a novel compound, 4-(2,3,5,6-tetramethylpyrazine-1)-4’-demethylepipodophyllotoxin. Appl Environ Microbiol. 2011;77(9):3023–3034. doi:10.1128/AEM.03047-10
6. Yuan Y, Wang YZ, Xu R, et al. Application of ionic liquids in the microwave-assisted extraction of podophyllotoxin from Chinese herbal medicine. Analyst. 2011;136(11):2294–2305. doi:10.1039/c0an00864h
7. Zhou JT, Jiang XY, He SY, et al. Rational design of multitarget-directed ligands: strategies and emerging paradigms. J Med Chem. 2019;62(20):8881–8914. doi:10.1021/acs.jmedchem.9b00017
8. Zhang WL, Pei JF, Lai LH. Computational multitarget drug design. J Chem Inf Model. 2017;57(3):403–412. doi:10.1021/acs.jcim.6b00491
9. Korcsmáros T, Szalay MS, Böde C, et al. How to design multi-target drugs. Expert Opin Drug Discov. 2007;2(6):799–808.
10. Malik V, Dhanjal JK, Kumari A, et al. Function and structure-based screening of compounds, peptides and proteins to identify drug candidates. Methods. 2017;131:10–21. doi:10.1016/j.ymeth.2017.08.010
11. Mulabagal V, Calderón AI. Development of an ultrafiltration-liquid chromatography/mass spectrometry (UF-LC/MS) based ligand-binding assay and an LC/MS based functional assay for Mycobacterium tuberculosis shikimate kinase. Anal Chem. 2010;82(9):3616–3621. doi:10.1021/ac902849g
12. Liang XX, Wu Q, Luan SX, et al. A comprehensive review of topoisomerase inhibitors as anticancer agents in the past decade. Eur J Med Chem. 2019;171:129–168. doi:10.1016/j.ejmech.2019.03.034
13. Sinha BK. Topoisomerase inhibitors. A review of their therapeutic potential in cancer. Drugs. 1995;49(1):11–19. doi:10.2165/00003495-199549010-00002
14. Delgado JL, Hsieh CM, Chan NL, et al. Topoisomerases as anticancer targets. Biochem J. 2018;475(2):373–398. doi:10.1042/BCJ20160583
15. Goradel NH, Najafi M, Salehi E, et al. Cyclooxygenase-2 in cancer: a review. J Cell Physiol. 2019;234(5):5683–5699. doi:10.1002/jcp.27411
16. Xu W, Huang YP, Zhang T, et al. Cyclooxygenase-2 gene polymorphisms and susceptibility to hepatocellular carcinoma: a meta-analysis based on 10 case-control studies. J Cancer Res Ther. 2018;14(8):S105–S113. doi:10.4103/0973-1482.172110
17. Mahboubi Rabbani SMI, Zarghi A. Selective COX-2 inhibitors as anticancer agents: a patent review (2014–2018). Expert Opin Ther Pat. 2019;29(6):407–427. doi:10.1080/13543776.2019.1623880
18. Gheblawi M, Wang K, Viveiros A, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126(10):1456–1474. doi:10.1161/CIRCRESAHA.120.317015
19. Hamming I, Cooper ME, Haagmans BL, et al. The emerging role of ACE2 in physiology and disease. J Pathol. 2007;212(1):1–11. doi:10.1002/path.2162
20. Luo LX, Qiu Q, Huang FF, et al. Drug repurposing against coronavirus disease 2019 (COVID-19): a review. J Pharm Anal. 2021;11(6):683–690. doi:10.1016/j.jpha.2021.09.001
21. Li YW, Zhou W, Yang L, et al. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol Res. 2020;157:104833. doi:10.1016/j.phrs.2020.104833
22. Kuba K, Imai Y, Ohto-Nakanishi T, et al. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. 2010;128(1):119–128. doi:10.1016/j.pharmthera.2010.06.003
23. Xie L, Lee DY, Shang Y, et al. Characterization of spirostanol glycosides and furostanol glycosides from anemarrhenae rhizoma as dual targeted inhibitors of 5-lipoxygenase and cyclooxygenase-2 by employing a combination of affinity ultrafiltration and HPLC/MS. Phytomedicine. 2020;77:153284. doi:10.1016/j.phymed.2020.153284
24. Qin SS, Ren YR, Fu X, et al. Multiple ligand detection and affinity measurement by ultrafiltration and mass spectrometry analysis applied to fragment mixture screening. Anal Chim Acta. 2015;886:98–106. doi:10.1016/j.aca.2015.06.017
25. Choi Y, Jermihov K, Nam SJ, et al. Screening natural products for inhibitors of quinone reductase-2 using ultrafiltration LC-MS. Anal Chem. 2011;83(3):1048–1052. doi:10.1021/ac1028424
26. Chen GL, Xu YB, Wu JL, et al. Hypoglycemic and hypolipidemic effects of Moringa oleifera leaves and their functional chemical constituents. Food Chem. 2020;333:127478. doi:10.1016/j.foodchem.2020.127478
27. Xu YB, Chen GL, Guo MQ. Potential anti-aging components from Moringa oleifera leaves explored by affinity ultrafiltration with multiple drug targets. Front Nutr. 2022;9:854882. doi:10.3389/fnut.2022.854882
28. Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1(3):1112–1116. doi:10.1038/nprot.2006.179
29. Feng HX, Chen GL, Zhang YL, et al. Potential multiple bioactive components from Sinopodophyllum hexandrum explored by affinity ultrafiltration with four drug targets. Phytomed Plus. 2021;2:100219. doi:10.1016/j.phyplu.2022.100219
30. Shen MR, He Y, Shi SM. Development of chromatographic technologies for the quality control of traditional Chinese medicine in the Chinese pharmacopoeia. J Pharm Anal. 2021;11(2):155–162. doi:10.1016/j.jpha.2020.11.008
31. Liu YQ, He GH, Li HL, et al. Plasma pharmacokinetics and tissue distribution study of roemerine in rats by liquid chromatography with tandem mass spectrometry (LC-MS/MS). J Chromatogr B Analyt Technol Biomed Life Sci. 2014;969:249–255. doi:10.1016/j.jchromb.2014.08.031
32. Ghani NA, Ismail NH, Asakawa Y. Constituents of fermented male flowers of Alnus sieboldiana (Betulaceae). Nat Prod Commun. 2017;12(1):57–58.
33. Yang Y, Chen Y, Zhong ZY, et al. Validated LC-MS/MS assay for quantitative determination of deoxypodophyllotoxin in rat plasma and its application in pharmacokinetic study. J Pharm Biomed Anal. 2014;88:410–415. doi:10.1016/j.jpba.2013.09.027
34. Yang Z, Wu YQ, Wu SH. A combination strategy for extraction and isolation of multi-component natural products by systematic two-phase solvent extraction-(13) C nuclear magnetic resonance pattern recognition and following conical counter-current chromatography separation: podophyllotoxins and flavonoids from Dysosma versipellis (Hance) as examples. J Chromatogr A. 2016;1431:184–196. doi:10.1016/j.chroma.2015.12.074
35. Yao CX, Na N, Huang LY, et al. High-throughput detection of drugs binding to proteins using desorption electrospray ionization mass spectrometry. Anal Chim Acta. 2013;794:60–66. doi:10.1016/j.aca.2013.07.016
36. Xie LW, Fu QC, Shi SY, et al. Rapid and comprehensive profiling of α-glucosidase inhibitors in Buddleja Flos by ultrafiltration HPLC-QTOF-MS/MS with diagnostic ions filtering strategy. Food Chem. 2021;344:128651. doi:10.1016/j.foodchem.2020.128651
37. Chen GL, Guo MQ. Screening for natural inhibitors of topoisomerases I from Rhamnus davurica by affinity ultrafiltration and high-performance liquid chromatography-mass spectrometry. Front Plant Sci. 2017;8:1521. doi:10.3389/fpls.2017.01521
38. Chen GL, Wu JL, Li N, et al. Screening for anti-proliferative and anti-inflammatory components from Rhamnus davurica Pall. using bio-affinity ultrafiltration with multiple drug targets. Anal Bioanal Chem. 2018;410(15):3587–3595. doi:10.1007/s00216-018-0953-6
39. Xiao JQ, Gao MX, Sun Z, et al. Recent advances of podophyllotoxin/epipodophyllotoxin hybrids in anticancer activity, mode of action, and structure-activity relationship: an update (2010–2020). Eur J Med Chem. 2020;208:112830. doi:10.1016/j.ejmech.2020.112830
40. Tabassum S, Zaki M, Afzal M, et al. New modulated design and synthesis of quercetin-CuII/ZnII-Sn2IV scaffold as anticancer agents: in vitro DNA binding profile, DNA cleavage pathway and Topo-I activity. Dalton Trans. 2013;42(27):10029–10041. doi:10.1039/c3dt50646k
41. Tang Q, Ji FL, Wang JY, et al. Quercetin exerts synergetic anti-cancer activity with 10-hydroxy camptothecin. Eur J Pharm Sci. 2017;109:223–232. doi:10.1016/j.ejps.2017.08.013
42. Zhang ZH, Chen S, Mei H, et al. Ginkgo biloba leaf extract induces DNA damage by inhibiting topoisomerase II activity in human hepatic cells. Sci Rep. 2015;5(1):14633. doi:10.1038/srep14633
43. Gowrishankar S, Muthumanickam S, Kamaladevi A, et al. Promising phytochemicals of traditional Indian herbal steam inhalation therapy to combat COVID-19 - an in silico study. Food Chem Toxicol. 2021;148:111966. doi:10.1016/j.fct.2020.111966
44. Guler HI, Tatar G, Yildiz O, et al. Investigation of potential inhibitor properties of ethanolic propolis extracts against ACE-II receptors for COVID-19 treatment by molecular docking study. Arch Microbiol. 2021;203(6):3557–3564. doi:10.1007/s00203-021-02351-1
45. Colunga Biancatelli RML, Berrill M, Catravas JD, et al. Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19). Front Immunol. 2020;11:1451. doi:10.3389/fimmu.2020.01451
46. Derosa G, Maffioli P, D’Angelo A, et al. A role for quercetin in coronavirus disease 2019 (COVID-19). Phytother Res. 2021;35(3):1230–1236. doi:10.1002/ptr.6887
47. Chen XQ, Wang ZX, Yang ZY, et al. Houttuynia cordata blocks HSV infection through inhibition of NF-κB activation. Antiviral Res. 2011;92(2):341–345. doi:10.1016/j.antiviral.2011.09.005
48. Cheng SC, Huang WC, Pang JHS, et al. Quercetin inhibits the production of IL-1β-induced inflammatory cytokines and chemokines in ARPE-19 cells via the MAPK and NF-κB signaling pathways. Int J Mol Sci. 2019;20(12):2957. doi:10.3390/ijms20122957
49. Fanunza E, Iampietro M, Distinto S, et al. Quercetin blocks Ebola virus infection by counteracting the VP24 interferon-inhibitory function. Antimicrob Agents Chemother. 2020;64(7):e00530–20. doi:10.1128/AAC.00530-20
50. Russo M, Moccia S, Spagnuolo C, et al. Roles of flavonoids against coronavirus infection. Chem Biol Interact. 2020;328:109211. doi:10.1016/j.cbi.2020.109211
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.