Back to Journals » OncoTargets and Therapy » Volume 12
Positive feedback loop of lncRNA HOXC-AS2/miR-876-5p/ZEB1 to regulate EMT in glioma
Authors Dong N, Guo J, Han S, Bao L, Diao Y, Lin Z
Received 20 May 2019
Accepted for publication 28 August 2019
Published 16 September 2019 Volume 2019:12 Pages 7601—7609
DOI https://doi.org/10.2147/OTT.S216134
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Nan Dong,1 Junxiu Guo,2 Song Han,1 Long Bao,3 Yi Diao,4 Zhixiong Lin1
1Department of Neurosurgery, Beijing Sanbo Brain Hospital, Capital Medical University, Beijing 100093, People’s Republic of China; 2Department of Neurosurgery, Shanxi Children’s Hospital, Taiyuan 030013, People’s Republic of China; 3Department of Neurosurgery, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, Liaoning 121000, People’s Republic of China; 4Department of Neurosurgery, Xuzhou Central Hospital, Xuzhou, Jiangsu 221009, People’s Republic of China
Correspondence: Zhixiong Lin
Department of Neurosurgery, Beijing Sanbo Brain Hospital, Capital Medical University, 50 Xiangshanyikesong Road, Beijing 100093, People’s Republic of China
Tel/Fax +86 010 62856707
Email [email protected]
Yi Diao
Department of Neurosurgery, Xuzhou Central Hospital, Xuzhou, Jiangsu 221009, People’s Republic of China
Tel/Fax+86 516 8395 6768
Email [email protected]
Purpose: Growing evidence has valued the diagnostic and therapeutic ability of long non-coding RNAs (lncRNAs) in various human tumors including glioma. Here, we investigated the biological function and potential mechanism of a novel cancer-related lncRNA, HOXC-AS2, in glioma.
Materials and methods: The expression of lncHOXC-AS2 was detected using qRT-PCR in glioma cells and tissues. A series of in vitro studies were performed to analyze the biological function of lncHOXC-AS2. Dual-luciferase reporter, RIP was used to determine the relation between lncHOXC-AS2, miR-876-5p and ZEB1. CHIP assay was performed to investigate the transcriptional regulation of HOXC-AS2.
Results: We found HOXC-AS2 was upregulated in glioma cells and tissues. Depletion of HOXC-AS2 was associated with the inhibition of migration, invasion and EMT process in glioma cells. Mechanism, HOXC-AS2 can sponge miR-876-5p to affect ZEB1 expression. Meanwhile, ZEB1 can bind promoter region of HOXC-AS2 and regulate HOXC-AS2 at transcriptional level.
Conclusion: Our results conclude that HOXC-AS2/miR-876-5p/ZEB1 constitutes a positive feedback loop to regulate EMT in GBM, providing a potential therapeutic target for glioma.
Keywords: HOXC-AS2, epithelial–mesenchymal transition, competing endogenous RNA, glioma
Introduction
Glioblastoma mulitiforme (GBM), the most malignant form of glioma, is one of the most aggressive and lethal human tumor in adults.1,2 Despite the improvement of multimodal therapeutic strategy, the range of median overall survival of GBM patients remains at 12–15 months.3 Therefore, it is necessary to clarify the molecular mechanism contribute to strong invasiveness of glioma and exploit effective-targeted therapies.
Epithelial–mesenchymal transition (EMT) causes cell to lose their epithelial characteristics, acquire migratory and invasive ability and away from epithelial cell community to become mesenchymal cells.4,5 Based on this theory, EMT has been recognized to occur in tumor progression,6 including prostate cancer,7 gastric cancer8 and breast cancer.9 Tumor cells and matrix components collaborative participate in the malignant progression and recurrence of glioma.10 It is precisely because of this, researchers of glioma EMT are essential to reverse malignant progression and reduce recurrence.
Non-coding RNAs (ncRNAs) are a group of non-protein coding transcripts, including long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs). Long non-coding RNAs are a class of non-coding RNA with more than 200 base pairs in length11 and closely relate to the malignant progression of multiple human tumors, including lung cancer,12 gallbladder cancer13 and glioma.14 Although plenty of lncRNA has been annotated, we still need to discover more tumor biomarkers and further discuss their contribution to glioma tumorigenesis. LncRNA HOXC cluster antisense RNA 2 (lncHOXC-AS2), located on chromosome 12q13.13, is a novel lncRNA for tumor, especially glioma. Thus, it is essential to clarify the biological function and potential molecular mechanism of HOXC-AS2 on glioma malignant progression.
Our current research, for the first time, reported that HOXC-AS2 function as an oncogene in glioma. Mechanistically, HOXC-AS2 indirectly regulated ZEB1 expression by sponging miR-876-5p. More importantly, ZEB1 can in turn up-regulated HOXC-AS2 via binding its promoter region. Our data confirm the existence of HOXC-AS2/miR-876-5p/ZEB1 positive feedback loop, provide theoretical basis for precision medicine of glioma.
Materials and methods
Glioma tissues and cell lines
LncRNA expression and survival data in glioma were downloaded from The Cancer Genome Atlas (TCGA) dataset (http://cancergenome.nih.gov). All glioma specimens and cerebral trauma samples (non-neoplastic brain tissues, NBTs) were obtained from Department of Neurosurgery, Beijing Sanbo Brain Hospital. The study was approved by the Ethics Committee of Capital Medical University and all patients were asked to write informed consent and the research was conducted in accordance with the Declaration of Helsinki. Human glioma cell lines (U87, LN229, U251, T98 and U118) and normal human astrocytes (NHAs) were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). All glioma cells were cultured with Dulbecco,s modified Eagle,s medium (DMEM) medium containing 10% fetal bovine serum (FBS), 1% penicillin and 1% streptomycin. NHAs were cultured with astrocyte medium (Carlsbad, CA, USA). All cells were maintained at 37°C with 5% CO2.
Cell transfection
HOXC-AS2 small interfering RNA (siHOXC-AS2) and control siRNA (siCtrl), ZEB1 small interfering RNA (siZEB1) and control siRNA (si-NC), were obtained from Genechem (Shanghai, China). And miR-876-5p mimics (miR-876-5p), miR-876-5p mimics control (miR-NC), miR-876-5p inhibitor (anti-miR-876-5p) were purchased from RiboBio (Guangzhou, China). The ORF region of ZEB1 cDNA was insert into pcDNA3.1 plasmid (ZEB1). RiboFECT CP Transfection Kit (RiboBio, Guangzhou, China) was purchased for transient transfection according to manufacturer’s instructions.
RNA extraction and qRT-PCR assay
We extracted total RNA from glioma tissues and cultured cells by using Trizol (Invitrogen, Carlsbad, USA) according to manufacture’s instructions. Extracted RNA was reverse transcribed into cDNA by using the PrimeScript RT reagent Kit (TaKara, Nanjing, China). Then, by using SYBR Premix ExTaq (Takara), quantitative real-time PCR (qRT-PCR) was performed and the results were normalized with U6. PCR 7300 real-time PCR system (Applied Biosystems, Foster City, USA) was chosen to carry out qRT-PCR analysis. All primer for lncRNA (HOXC-AS2), miRNA (miR-876-5p) and internal control (U6) were purchased from Ribobio (Guangzhou, China). All primer sequences were listed in Table S1. The results were analyzed by 2-△△Ct method.
Western blot assay
Western blot assay was performed according to our previous study.15 Antibody against ZEB1, Vimentin, N-cadherin and β-actin were purchased from Cell Signaling Abcam (Cambridge, UK).
Wound healing assay and transwell assay
Wound healing assay and transwell assay were performed as previously described.15 Briefly, for transwell assay, glioma cells were seeded in transwell chambers (matrigel coating) which containing serum-free medium. Then, the transwell chambers were placed in 24-well plate containing complete medium and glioma cells were incubated for 24 h.
Luciferase assay
Luciferase plasmid containing wild type (WT) or mutant (MUT) HOXC-AS2 or ZEB1 sequences were constructed by Genechem (Shanghai, China). Briefly, the complementary DNA fragment containing the wild type or mutant HOXC-AS2 and 3ʹ untranslated region (UTR) of ZEB1 was sub-cloned downstream of the luciferase gene within the pGL3-Basic luciferase reporter vector (Promega). Mimics control or miR-876-5p mimics were co-transfected with luciferase plasmid into LN229 and U251 cells. After 48 h transfection, the luciferase assays were analyzed using Dual-Luciferase Kit (Promega). Dual-Luciferase Reporter Assay System (Promega) was chosen to measure luciferase activities.
RIP assay
Briefly, magnetic beads were pre-conjugated with antibody against Argonaute2 (Ago2). Related LN229 and U251 glioma cell lysates were harvested and treated with bead-antibody complex. Normal mouse IgG was used as a negative control. RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was chosen for RIP and all experiments were performed according to manufacturer’s instructions.
CHIP assay
Chip assay was performed using Chromatin Immunoprecipitation (CHIP) Kit (Millipore, Massachusetts, USA) according to the manufacturer’s instructions. Briefly, the chromatin of the LN229 and U251 cells was treated with 3 μg specific ZEB1 antibody. Immunoprecipitated DNA was analyzed by RT-PCR.
Statistical analysis
Graphpad Prism 7 (La Jolla, CA, USA) and SPSS 22.0 software (IBM, Corp., Armonk, New York, USA) were chosen to analyze statistical analysis. All value was expressed as mean ± Standard Error of Mean (SEM). Student’s t-test or ANOVA were performed to evaluate data. Differences were judged to be statistically significant at P<0.05.
Results
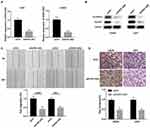
HOXC-AS2 is upregulated in glioma tissues and cells
We first used TCGA database to analyze the differential expression of lncRNAs in glioma (Figure 1A). Combined with survival analysis, HOXC-AS2 was chosen for further study because of its significant difference in both expression and survival (Figure 1A, B and Table S2). By using qRT-PCR, we found that high-grade glioma (HGG) tissues had higher HOXC-AS2 level than low-grade glioma (LGG) (Figure 1C). Then, we detect the expression level of HOXC-AS2 in glioma cells. Almost all glioma cells were found to highly express HOXC-AS2 in comparison with normal human astrocytes NHAs (Figure 1D). Furthermore, we performed fluorescence in situ hybridization (FISH) assays to analyzed HOXC-AS2 level in different grades of glioma (Figure 1E, Figure S1A and Table S3). Altogether, our data indicated that HOXC-AS2 is enriched in glioma cells and tissues, suggesting HOXC-AS2 has the ability to become a prognostic biomarker.
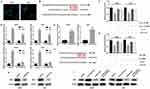
Knockdown of HOXC-AS2 suppresses migration, invasion and EMT program in glioma cells
To further confirm the effect of HOXC-AS2 on glioma progress, HOXC-AS2 was knockdown by using small interfering RNA (siRNA) in LN229 and U251 cells which express high level of HOXC-AS2 than other glioma cells (Figure 1D). Low level of HOXC-AS2 was found in LN229 and U251 cells after transfected with siHOXC-AS2 (Figure 2A). Due to the depletion of the structure of epithelial cells, E-cadherin is hardly expressed in gliomas compared to other epithelial-type tumors. Thus, we paid great attention to the expression change of mesenchymal phenotype markers, Vimentin and N-cadherin, which can reflect the EMT program in glioma. By using Western blot assay, we found that knockdown of HOXC-AS2 means the decrease of Vimentin and N-cadherin (Figure 2B). EMT program is usually followed by the changes of cell migration and invasion. In order to confirm our conjecture, we performed wound healing and transwell assays to detect the migratory and invasive ability of glioma cells. As shown in Figure 2C and D, depletion of HOXC-AS2 leads to delayed wound healing and decreased invasive ability. These results showed that HOXC-AS2 inhibition can reduce EMT progression and malignancy in glioma.
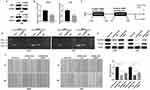
HOXC-AS2 indirectly regulated ZEB1 via sponging miR-876-5p
Increasing evidence demonstrates that lncRNAs can function as competing endogenous RNAs (ceRNAs) for miRNAs to regulate target gene expression.16,17 The mechanism by which lncRNA works is mainly due to its location. Therefore, we first analyzed the distribution of HOXC-AS2 by using FISH analysis. The results showed that HOXC-AS2 mainly enriched in the cytoplasm (Figure 3A), indicating that HOXC-AS2 might function as a ceRNA for miRNAs. By using DIANA database (http://diana.imis.athena-innovation.gr/), we predicted potential target miRNA for HOXC-AS2 and we focus on a miR-876-5p which had been considered as a cancer suppressor gene in osteosarcoma18 and hepatocellular carcinoma.19 Then, we analyzed the expression of miR-876-5p in CGGA database and the result showed that miR-876-5p was downregulated in glioma (Figure S1C), consistent with previous reports.20 To confirm our prediction, we performed a dual luciferase reporter assay to confirm that miR-876-5p binds directly to HOXC-AS2. The observed miR-876-5p-mediated decreased of luciferase activity was limited by mutation (MUT) of HOXC-AS2 (Figure 3B and C). We used RNA-binding protein immunoprecipitation (RIP) assay to better characterize the relation between HOXC-AS2 and miR-876-5p. Compared with IgG precipitates, the enrichment of HOXC-AS2 and miR-876-5p immunoprecipitated with Ago2 was much higher in control glioma cells. Besides, depletion of miR-876-5p decreased enrichment of HOXC-AS2 and miR-876-5p in Ago2 precipitates (Figure 3D). Meanwhile, knockdown of HOXC-AS2 conducted the increase of miR-876-5p (Figure 3E). Based on ceRNA hypothesis, we used bioinformatic website (Targetscan, http://www.targetscan.org/) and found that ZEB1 is a potential target of miR-876-5p. To confirm our prediction, we performed dual-luciferase reporter assay and observed luciferase activity in LN229 and U251 cells that were obviously reduced after co-transfected with ZEB1 wild-type (WT) vector and miR-876-5p mimics (Figure 3F and G). Meanwhile, after miR-876-5p mimics infection in LN229 and U251 cells, ZEB1 protein level was significantly decreased (Figure 3H). Then, we co-transfected HOXC-AS2 and miR-876-5p inhibitor into glioma cells, Western blot assay showed that HOXC-AS2 significantly reduced ZEB1 level but this reduction was rescued by the miR-876-5p inhibitor (Figure 3I). All in all, our results confirmed that HOXC-AS2 positively regulated ZEB1 via sponging miR-876-5p, consisting of HOXC-AS2/miR-876-5p/ZEB1 pathway.
HOXC-AS2 and ZEB1 form a positive feedback loop
Recent studies have shown that transcription factors can regulate lncRNA expression via directly bind to specific regions of promoters.21 Thus, we considered whether ZEB1, a well-known EMT-related transcription factor, could affect HOXC-AS2 expression. To test our conjecture, we used JASPAR database (http://jaspar.binf.ku.dk/) and found that ZEB1 has potential binding sites in HOXC-AS2 promoter region. For further confirmation, we knockdown ZEB1 in LN229 and U251 cells (Figure 4A), and analyzed HOXC-AS2 expression level by using qRT-PCR. The results showed a positive correlation between HOXC-AS2 and ZEB1 levels (Figure 4B). By scanning the promoter region of HOXC-AS2 with JASPAR database, two putative ZEB1 binding sites with highest scores were chosen (Figure 4C). We performed chromatin immunoprecipitation assay (CHIP) to elucidate the binding of ZEB1 and HOXC-AS2 promoter region. Only Site 2 group had the PCR products in the ZEB1 immunoprecipitation, indicating that ZEB1 bound to HOXC-AS2 promoter at the −1375–−1327 bp region (Figure 4D). Together, these results showed that ZEB1 can regulate HOXC-AS2 at transcription level, indicating the existence of HOXC-AS2/miR-876-5p/ZEB1 positive feedback loop.
To further investigate the synergistic regulation of ZEB1 and HOXC-AS2 on glioma EMT progress, we performed Western blot assay and found that Vimentin, N-cadherin and ZEB1 expression were down-regulated in the wake of HOXC-AS2 knockdown (Figure 4E). In addition, the down-regulation could be reversed by ZEB1 over-expression (Figure 4E). The observations were verified by wound healing and transwell assays (Figure 4F and Figure S1B). Altogether, we concluded that HOXC-AS2/miR-876-5p/ZEB1 positive feedback loop contributes to EMT progress in glioma.
Discussion
Over the past decades, the research on long non-coding RNAs (lncRNA) have been widely developed. In 2011, Salmena, L and his colleagues first proposed the ceRNA hypothesis.22 Based on this theory and our experimental data, we confirm the presence of HOXC-AS2/miR-876-5p/ZEB1 positive feedback loop which has a vital role in glioma malignant progress.
More than 90% human genome is transcribed, but there are only few genes can encode protein, less than 3%. As a non-coding RNA, lncRNA has been studied as a key factor of multiple human diseases, including cancers.23 Several studies have focused on the biological function of lncRNAs on malignant behaviour, such as proliferation,13 migration,24 invasion,25 angiogenesis26 and chemoresistance.27 Epithelial–mesenchymal transition (EMT), a key factor of tumor invasion enhancement, has been proved to be involved in malignant progress of glioma.10,28 Our present study, for the first time, reported a novel lncRNA lncHOXC-AS2 (ENSG00000250133) which is located on chromosome 12q13.13, was up-regulated in glioma tissues and cells. Knockdown of HOXC-AS2 restrained invasive ability and reduced expression levels of N-cadharin and Vimentin. Thus, our results shed insight on the oncogenic function of HOXC-AS2 to promote EMT in glioma cells.
Competing endogenous RNAs (ceRNAs), a widespread interaction network, widely present in tumors. In brief, lncRNAs or circular RNAs (circRNAs) regulate their target microRNAs (miRNAs) via binding and titrating them off their binding sites on protein-coding messengers.22,29 Based on functional observations, we aimed to further analyze the molecular mechanism of HOXC-AS2. It is well-known that the mechanism of lncRNA is related to its intracellular localization. Thus, we first located HOXC-AS2 in glioma cells and found that HOXC-AS2 was enriched in the cytoplasm. Subsequent bioinformatics analysis combines with luciferase assays, RIP assays and Western blot assays verified that miR-876-5p was both target of HOXC-AS2 and ZEB1. Previously, Xue et al reported that miR-876-5p inhibits migration and invasion in glioma cells30 and our results are consistent with this conclusion of miR-876-5p in cell migration, invasion and EMT program.
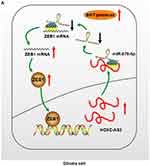
Zinc-finger E-box binding homeobox 1 (ZEB1) is a kind of zinc-finger protein which plays an important role in normal embryonic development.31 Recent studies focused on the relation between ZEB1 and tumor biological behavious, including metastasis,32 invasion,33 angiogenesis34 and chemoresistance.35 Moreover, ZEB1 acts as a transcription factor, can binding specific region of gene promoter.36 Here, we show that ZEB1 is indirectly regulated by HOXC-AS2 via sponging miR-876-5p. In turn, ZEB1 can bind promoter region of HOXC-AS2 and regulates its expression. Thus, we verify the presence of HOXC-AS2/miR-876-5p/ZEB1 positive feedback loop, which can enhance migrational and invasiveness ability as well as EMT program (Figure 5A). Our study provides evidence for the existence of a positive feedback loop between lncRNAs and their down-regulated targets, which may have a crucial effect on glioma development and progression.
Collectively, our present study highlights the importance of correlation between HOXC-AS2, miR-876-5p and ZEB1. HOXC-AS2 expression is upregulated in glioma tissues and cells. Increased expression of HOXC-AS2 can suppress miR-876-5p level, leads an up-regulation of ZEB1. Overexpressed ZEB1 then in turn up-regulates the expression of HOXC-AS2, which constitute a positive feedback loop and enhance migration, invasion and EMT program in glioma. Thus, the HOXC-AS2/miR-876-5p/ZEB1 feedback loop has potential to be a therapeutic target for glioma treatment.
Acknowledgment
This work was supported by a grant from the Neuro-Oncology Research Program of CSNO Grant (CSNO-2016-MSD01).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pekmezci M, Rice T, Molinaro AM, et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017;133(6):1001–1016. doi:10.1007/s00401-017-1690-1
2. Kotliarova S, Fine HA. SnapShot: glioblastoma multiforme. Cancer Cell. 2012;21(5):710–710.e711. doi:10.1016/j.ccr.2012.04.031
3. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA:A Cancer Journal for Clinicians. 2010; 60: 166–193.
4. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi:10.1172/JCI39104
5. Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi:10.1038/cr.2009.5
6. Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25(11):675–686. doi:10.1016/j.tcb.2015.07.012
7. Mulholland DJ, Kobayashi N, Ruscetti M, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–1889. doi:10.1158/0008-5472.CAN-11-3132
8. Yoo YA, Kang MH, Lee HJ, et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71(22):7061–7070. doi:10.1158/0008-5472.CAN-11-1338
9. Zhou W, Ye XL, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10(483):undefined. doi:10.1126/scisignal.aak9557
10. Karsy M, Gelbman M, Shah P, Balumbu O, Moy F, Arslan E. Established and emerging variants of glioblastoma multiforme: review of morphological and molecular features. Folia Neuropathol. 2012;50(4):301–321. doi:10.5114/fn.2012.32361
11. Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154(5):996–1009. doi:10.1016/j.cell.2013.07.047
12. Yu T, Zhao Y, Hu Z, et al. MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 2017;77(21):5782–5794. doi:10.1158/0008-5472.CAN-17-0671
13. XS W, Wang F, HF L, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18(10):1837–1853. doi:10.15252/embr.201744147
14. Liu X, Zheng J, Xue Y, et al. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8(4):1084–1105. doi:10.7150/thno.21740
15. Li Z, Guo J, Ma Y, Zhang L, Lin Z. Oncogenic role of microRNA-30b-5p in glioblastoma through targeting proline-rich transmembrane protein 2. Oncol Res. 2018;26(2):219–230. doi:10.3727/096504017X14944585873659
16. Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299–6310. doi:10.1158/0008-5472.CAN-16-0356
17. Zhang E, Han L, Yin D, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45(6):3086–3101. doi:10.1093/nar/gkw1247
18. Xie W, Xiao J, Wang T, Zhang D, Li Z. MicroRNA-876-5p inhibits cell proliferation, migration and invasion by targeting c-Met in osteosarcoma. J Cell Mol Med. 2019;23(5):3293–3301. doi:10.1111/jcmm.2019.23.issue-5
19. Wang Y, Xie Y, Li X, et al. MiR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol Res Pract. 2018;214(7):1024–1030. doi:10.1016/j.prp.2018.04.012
20. Giunti L, Da Ros M, De Gregorio V, et al. A microRNA profile of pediatric glioblastoma: the role of NUCKS1 upregulation. Mol Clin Oncol. 2019;10(3):331–338.
21. Kültz D, Csonka L. What sets the TonE during osmotic stress? Proc Natl Acad Sci USA. 1999;96(5):1814–1816. doi:10.1073/pnas.96.5.1814
22. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi P. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
23. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi:10.1016/j.tcb.2011.04.001
24. Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105. doi:10.1186/s12943-018-0849-2
25. Liu L, Meng T, Yang XH, et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22(2):283–299. doi:10.3233/CBM-171011
26. Zhu Y, Zhang X, Qi L, et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7(12):14429–14440.
27. Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9(7):742. doi:10.1038/s41419-018-0793-5
28. Siebzehnrubl FA, Silver DJ, Tugertimur B, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. doi:10.1002/emmm.201302827
29. Jalali S, Bhartiya D, Lalwani M, Sivasubbu S, Scaria V, Zhang L. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8(2):e53823. doi:10.1371/journal.pone.0053823
30. Wang Z, Xue Y, Wang P, Zhu J, Ma J. MiR-608 inhibits the migration and invasion of glioma stem cells by targeting macrophage migration inhibitory factor. Oncol Rep. 2016;35(5):2733–2742. doi:10.3892/or.2016.4652
31. Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi:10.1038/ncb1722
32. Li L, Zheng YL, Jiang C, et al. HN1L-mediated transcriptional axis AP-2γ/METTL13/TCF3-ZEB1 drives tumor growth and metastasis in hepatocellular carcinoma. Cell Death Differ. 2019.
33. Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi:10.1038/embor.2008.74
34. Liu L, Tong Q, Liu S, et al. ZEB1 upregulates VEGF expression and stimulates angiogenesis in breast cancer. PLoS One. 2016;11(2):e0148774. doi:10.1371/journal.pone.0148774
35. Cui Y, Qin L, Tian D, et al. ZEB1 promotes chemoresistance to cisplatin in ovarian cancer cells by suppressing SLC3A2. Chemotherapy. 2018;63(5):262–271. doi:10.1159/000493864
36. Yu H, Zheng J, Liu X, et al. Transcription factor NFAT5 promotes glioblastoma cell-driven angiogenesis via SBF2-AS1/miR-338-3p-Mediated EGFL7 expression change. Front Mol Neurosci. 2017;10:301. doi:10.3389/fnmol.2017.00301
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.