Back to Journals » OncoTargets and Therapy » Volume 9
Positive expression of KIF20A indicates poor prognosis of glioma patients
Authors Duan J, Huang W, Shi H
Received 28 June 2016
Accepted for publication 30 August 2016
Published 2 November 2016 Volume 2016:9 Pages 6741—6749
DOI https://doi.org/10.2147/OTT.S115974
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Jia Duan,* Wei Huang,* Haiping Shi
Department of Neurosurgery, Suining Central Hospital, Suining, Sichuan, People’s Republic of China
*These authors contributed equally to this work
Abstract: Glioma patients have a poor overall survival; however, patients can show distinct clinical outcomes due to the high heterogeneity of the tumor, which may be indicated by certain clinicobiological parameters. Kinesin family member 20A (KIF20A), which participates in cytokinesis and intracellular transportation, has been recently reported to be upregulated in pancreatic cancer, breast cancer, and bladder cancer. In the current study, we investigated the expression of KIF20A in gliomas and its significance in predicting the prognosis after surgery. We found that KIF20A positive expression in glioma tissues correlated significantly with Ki67 protein expression and advanced World Health Organization grade. Univariate and multivariate analysis revealed that KIF20A can act as an independent prognostic factor for predicting the overall survival of glioma patients. Moreover, we demonstrated that KIF20A can positively regulate the expression of Ki67 in glioma cell lines. Correspondingly, overexpression of KIF20A can promote cell proliferation and invasion, whereas knockdown of KIF20A can inhibit cell viability and invasion capacity. In vitro study also showed that under the treatment of plumbagin, an anticancer drug, KIF20A expression decreased in a dose-dependent manner. In addition, the overexpression of KIF20A can also increase the drug resistance toward plumbagin, which provided the possibility that KIF20A may contribute to the chemotherapy resistance of gliomas.
Keywords: glioma, invasion, KIF20A, prognosis, proliferation
Introduction
Gliomas account for ~80% of primary malignant neoplasms of the central nervous system and represent the most aggressive primary brain tumors in adults. Gliomas can be classified into grade II, grade III, and grade IV based on the 2007 World Health Organization (WHO) Classification of Tumors of the Central Nervous System.1 Although great improvements in surgery, radiation, and chemotherapies have been made during the past decades, the median survival for patients with glioblastoma (WHO grade IV), the most aggressive glioma, is <15 months as a result of rapid proliferation, invasive growth, and chemotherapy resistance.2 Nevertheless, the survival time can vary among patients due to the heterogeneity of the tumor. For example, it can range from months up to 5 years for glioblastoma patients.3 Given that glioma cells show heterogeneous molecular expression,4 the current diagnosis methods based on morphology can be limited for prognosis evaluation. Hence, it is still under critical demand to identify novel biomarkers that may help in developing patient-specific treatments as well as improving the prognosis of glioma patients.
Kinesins (KIFs) are a superfamily of molecular motors that participate in cell mitosis,5,6 migration, and intracellular transport through unidirectional travel along microtubules.7,8 Kinesin family member 20A (KIF20A), also known as RAB6KIFL, plays an important role in cytokinesis.9,10 It was first identified to function in the dynamics of the Golgi apparatus through interaction with the Rab6A/B GTPase. Moreover, it has been reported that KIF20A is overexpressed in melanoma,11 bladder cancer,10,12 breast cancer,13–15 and pancreatic cancer.16 Knockdown of KIF20A in pancreatic cancer cells can significantly inhibit cell proliferation,17,18 indicating the significance of its overexpression in human cancers. Studies have also reported that several peptides derived from KIF20A can be helpful to activate the immune system and kill the cancer cells which endogenously express KIF20A antigen.16,19,20 Several groups are carrying out the phase I/II clinical trials for immunotherapy using these KIF20-derived peptides.21 However, it is unclear whether KIF20A is aberrantly expressed in glioma or whether it plays a role in glioma cell proliferation and survival.
In the current study, we explored the expression of KIF20A in clinical glioma tissues and demonstrated that KIF20A positive expression in gliomas can indicate poorer clinical outcome through univariate and multivariate analysis. In addition, we performed the knockdown and overexpression experiment in glioma cell line U87 and A172, respectively. Through in vitro study, we identified the role of KIF20A in regulating glioma cell proliferation, invasion, and drug resistance toward plumbagin, an anticancer agent.22,23 To our knowledge, this is the first report investigating the expression and role of KIF20A in gliomas. Our results not only revealed that KIF20A is an independent prognostic factor of gliomas but also provided evidence for the potency of KIF20A being a target molecule for glioma therapy.
Patients
From March 2002 to October 2011, a total of 119 glioma patients who underwent surgery treatment at Suining Central Hospital (Sichuan, People’s Republic of China) were retrospectively enrolled into this study. The formalin-fixed and paraffin-embedded resected or biopsy glioma tissues were collected. The histological tumor grade was determined using the WHO classification system.1 For prognosis analyses, the overall survival (OS) was defined as the interval between surgery and death or the last follow-up. This study was approved by the Ethics Committees of Suining Central Hospital, and written informed consent was obtained from each participant. Detailed clinicopathologic characteristics of all the patients are shown in Table 1.
Methods
Immunohistochemistry
For detection of KIF20A and Ki67 expression in glioma tissues, deparaffinized and rehydrated slides were treated in 10 mM citrate buffer (pH 6.0) at 95°C for antigen recovery, followed by quenching the endogenous peroxidase activity with 3% H2O2 incubation. Five percent fetal bovine serum (FBS) was used for blocking the nonspecific binding sites. Slides were then incubated with primary antibodies (anti-KIF20A, Abcam, Cambridge, UK, #ab104118; anti-Ki67, Abcam, #ab15580, 1:200) at 4°C overnight. DAB staining kit (Tiangen, People’s Republic of China) was used to detect the immunoreactivity according to the manufacturer’s instructions.
All slides were examined by two pathologists who had no prior knowledge of the clinical data. For the immunohistochemistry (IHC) evaluation, we regard the cells which were stained with yellow or brown color as positive-stained cells. Then we counted the cells under a light microscope and calculated the average percentage of positive cells in five random fields. If the percentage of positive cells was <25%, then we classified it as negative expression of KIF20A; if the percentage of positive cells was ≥25%, then we defined it as positive expression of KIF20A. Therefore, we sorted the patients into negative-expression group and positive-expression group and performed statistical analyses subsequently.
Cell culture and transfection
The glioma cell lines A172, U251, and U87 were obtained from the American Type Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS, 100 U/mL penicillin, and 0.1 mg/mL streptomycin at 37°C in 5% CO2.
The plasmid pcDNA3.0 vector was obtained from Thermo Fisher Scientific (Waltham, MA, USA). KIF20A cDNA sequence fragment with KpnI and XhoI cutting sites was generated by polymerase chain reaction (PCR) from human cDNA. The PCR products were then digested and subcloned into the pcDNA3.0 vector small interfering RNAs (siRNA) were chemically synthesized from Sigma-Aldrich Co. (St Louis, MO, USA) with the following sequences: Control-siRNA: 5′-UUCUCCGAACGUGUCACGUTT-3′, KIF20A-siRNA#1: 5′-GTTCTCAGCCATTGCTAGC-3′,17 KIF20A-siRNA#2: 5′-GCAGCAGGUUCCAUCUGAG-3′.
The transient transfections of pcDNA3.0 plasmids and siRNAs were performed with Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s instructions. The efficiency of transient transfection was assessed by Western blotting.
Cell lysis and Western blotting
Cells were washed with cold phosphate-buffered saline (PBS) and then homogenized using NP40 lysis buffer (Beyotime Biotechnology, Jiangsu, People’s Republic of China) containing protease inhibitors and phosphatase inhibitors. The total amount of proteins in cell lysates was quantified with Pierce™ bicinchoninic acid a protein assay kit (Thermo Fisher Scientific). After denatured with the sodium dodecyl sulfate (SDS) loading buffer and boiled for 5 min, ~30 μg proteins were loaded and separated by SDS-polyacrylamide gel electrophoresis and then electrotransferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked in 5% nonfat milk in Tris-buffered saline-Tween 20 followed by immunoblotting overnight at 4°C with primary antibodies (anti-KIF20A, Abcam, #ab104118; anti-Ki67, Abcam, #ab15580; anti-glyceraldehyde 3-phosphate dehydrogenase [GAPDH], Santa Cruz Biotechnology Inc., Dallas, TX, USA, sc-47724, 1:1,000). After being washed with Tris-buffered saline-Tween 20 and incubated with the secondary antibodies for another 1 h, the immunoreactivity was detected with the BeyoECL Plus reagent (Beyotime Biotechnology).
Cell proliferation assay and cell invasion assay
Cell viability was measured using the Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) assay following the manufacturer’s instructions. Briefly, transfected cells were seeded into 96-well plates at a density of 2×103 cells/well and cultured for 24, 48, 72, 96, and 120 h. The CCK-8 reagents were added into the wells at designated time points and incubated for another 2 h at 37°C. The absorbance was measured at 450 nm using the automated plate reader. Each experiment was performed in triplicate.
The invasion capacity of U87 and A172 cells was evaluated using the Boyden chamber Matrigel invasion assay (Cell Biolabs, San Diego, CA, USA). Briefly, 5×104 transfected cells were planted into the upper chamber and cultured with DMEM containing 1% FBS, while DMEM with 10% FBS was added into the lower chamber as the chemoattractant. Following 48 h incubation, noninvading cells were removed gently with a cotton swab, then fixed the cells that migrated to the lower surface of membrane with methanol, and stained with crystal violet. Cell counting was performed in five randomly selected fields for each well. The final invasion capacity was normalized by the protein amount in each well. Each experiment was performed in triplicate.
Reverse transcription and quantitative real time PCR (qRT-PCR)
Briefly, the total RNA was extracted from U87 cells with TRIZOL reagent solution (Thermo Fisher Scientific), and reverse DNA synthesis with SuperScript III reverse transcriptase (Thermo Fisher Scientific) was performed. The relative amount of KIF20A mRNA was evaluated by qRT-PCR with the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) and calculated with 2−ΔΔct method. The mRNA level of GAPDH was used for normalization.
The primer sequence for Kif20A detection was 5′-CAAGGGCCTAACCCTCAA-3′ and 5′-TCTGTCGTCTCTACCTCCCTAGA-3′,18 while the primer sequence for human GAPDH was 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′.24
Plumbagin resistance and half maximal inhibitory concentration (IC50) determination
The effect of KIF20A expression on plumbagin resistance of A172 glioma cells was assessed with the CCK-8 assay. Plumbagin has been reported to induce cell cycle arrest and cell apoptosis in various cancer cell lines including breast, lung, and glioma. However, the relationship between KIF20A and plumbagin resistance in glioma has not been studied.
Briefly, transfected cells (5×103) were seeded into a 96-well plate. After cultured for 24 h for cell adhesion, the cells were treated with plumbagin (Sigma-Aldrich Co.) with different concentrations ranged from 0.05 to 50 μM (0.5, 0.1, 0.2, 0.5, 1, 2, 5, 10, 25, and 50 μM). After 48 h of coincubation, CCK-8 reagent was added to each well. Approximately 2 h later, the absorbance at 450 nm (optical density [OD450]) was measured using an automated plate reader. The relative cell viability was calculated as (OD450 of mean plumbagin-treated − blank)/(OD450 of mean PBS-treated control − blank) ×100. The IC50 value was determined by the dose–response–inhibition curve using Graphpad Prism software 5.0.
Statistical analysis
All data was analyzed using Statistical Package for the Social Sciences 20.0 (IBM Corporation, Armonk, NY) or Graphpad Prism software 5.0. The correlation between KIF20A and Ki67 protein expression was determined using Pearson’s correlation test. Associations between clinicopathological parameters with KIF20A expression were identified using Pearson’s chi-square test. OS was evaluated by the Kaplan–Meier analysis and the difference between groups was assessed by log-rank test, while the Cox proportional hazards regression was performed to carry out the multivariate analysis. For the quantitative assays, the statistical significance was determined using Student’s t-test. P<0.05 was regarded as statistically significant.
Results
Patients characteristics and relationship with KIF20A expression
Of the 119 glioma patients recruited, there were 51 women (42.9%) and 68 men (57.1%) with an age range from 23 to 64 years old. The majority of the tumor size was <5 cm (71/119 cases, 59.7%). As for the histological classification, 29 cases (24.4%) suffered with WHO grade II, 48 cases with WHO grade III (40.3%), and the other 42 cases (35.3%) with WHO grade IV. Gross total resection was performed for 51 patients (42.9%), subtotal resection for 36 patients (30.3%), and the other 32 patients (26.9%) underwent partial resection or biopsy. We also evaluated the Karnofsky performance score25 for all of the patients before surgery treatment.
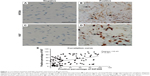
In the validation cohort, 63 cases (52.9%) were defined as KIF20A negative expression (Figure 1A) according to the IHC results, while the other 56 cases (47.1%) were identified as KIF20A positive expression which mainly existed in the cytoplasm of glioma cells (Figure 1B). We also investigated the expression of Ki67 protein in all of the glioma tissues (Figure 1C and D). Interestingly, we revealed the corresponding correlation between KIF20A expression and Ki67 expression (r=0.43, P<0.001, Figure 1E) through Pearson’s correlation test. Ki67 has been confirmed as a predictor for cell proliferation and malignant potential of cancers. The correlation between KIF20A and Ki67 expression indicates the carcinogenic role of KIF20A in gliomas.
Clinicopathologial features of the patients and their relationship with KIF20A expression were summarized in Table 1. These features include gender, age, tumor size, WHO grade, Karnofsky score, and surgery procedures. Significant correlation between WHO grade and KIF20A expression was identified. For the 29 patients with WHO grade II, nine cases (31.0%) were defined as KIF20 positive, while the KIF20A positive percentage for patients with WHO grades III and IV is 41.7% (20/48 cases) and 64.3% (27/42 cases), respectively. The correlation of KIF20A with Ki67 and WHO grade implied that this specific KIF may play roles in the tumorigenesis and development of glioma.
Positive expression of KIF20A is associated with poor prognosis of glioma patients
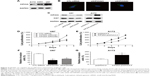
Kaplan–Meier survival analysis and log-rank test were used to evaluate the effects of various clinicopathological characteristics and KIF20A expression on OS of glioma patients (Table 2). The results demonstrated that patients with positive KIF20A expression showed poorer OS compared with the patients with negative KIF20 expression (median 16.0 vs 39.0 months, P=0.006; Figure 2A). Other conventional prognostic factors include WHO grade (P=0.002, Figure 2B) and surgery procedure (P=0.001, Figure 2C). No correlation was detected between OS and other clinicopathological features such as gender, age, tumor size, or Karnofsky score (Table 2).
Further analysis using the multivariate Cox proportional hazards model revealed that KIF20A expression (hazard ratio [HR] =1.614, 95% confidence intervals [CI]: 1.042–2.499, P=0.032), WHO grade (HR =1.510, 95% CI: 1.136–2.007, P=0.005), and surgery procedure (HR =1.628, 95% CI: 1.252–2.116, P<0.001) were all significantly correlated with OS (Table 3).
  | Table 3 Multivariate analysis for the prognostic factors of glioma patients |
Together these findings indicate that KIF20A expression can be a novel independent prognostic factor for the OS of glioma patients.
KIF20A expression can regulate the proliferation and invasion of glioma cells
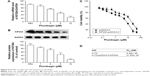
The expression pattern of KIF20A in glioma cells remains unclear, thus we assessed the expression of KIF20A protein in U251, U87, and A172 cell lines by Western blotting. Our results showed that KIF20A expression was significantly lower in the A172 cells than that in U251 and U87 cells, while U87 cells possess the highest KIF20A expression level (Figure 3A). Therefore, we chose the U87 cells to perform the siRNA knockdown experiment, while overexpressing KIF20A in the A172 cells. Corresponding to the IHC results, we also found that the expression level of Ki67 protein is positively regulated by the KIF20A (Figure 3B). Since Ki67 is a well-known cell proliferation biomarker and is correlated with the clinical course of cancer, our results indicated KIF20A may participate in the cell proliferation process of glioma cells. In addition, the expression of KIF20A was significantly associated with advanced WHO grade in human glioma tissues (Table 1), hence we further analyzed the effect of KIF20A on biological characteristics of glioma cell lines. We found that siRNA of KIF20A can inhibit cell proliferation and invasion (Figure 3C and E), while KIF20A-overexpression can upregulate proliferation and invasion capacity (Figure 3D and F).
The relationship between KIF20A expression and plumbagin resistance
Plumbagin is an anticarcinogenic drug which has been reported to be effective in inhibiting cell survival of lung cancer, pancreatic cancer, and glioma cells. We found that both the mRNA and protein expression level of KIF20A were downregulated by plumbagin treatment in a dose-dependent manner (Figure 4A and B) according to the RT-qPCR and Western blotting results. In addition, we investigated the effect of KIF20A expression on plumbagin resistance and demonstrated that KIF20A overexpression can significantly increase the IC50 of A172 cells toward plumbagin (8.45±1.26 vs 21.53±1.35 μM, P<0.05; Figure 4C and D).
Discussion
Glioma is one of the most lethal kinds of cancer. Its rapid proliferation and invasive growth makes the surgery resection very difficult and leads to high lethality. Identifying novel molecular biomarkers for glioma could be helpful for better understanding of the carcinogenic mechanisms and for development of more effective therapeutic treatments.26–28
KIF20A is a member of the KIF family, which has been reported to be upregulated in pancreatic cancer,29 breast cancer,14 and hepatocellular cancer.30 In the current study, we investigated the expression of KIF20A protein in gliomas using IHC and found that the expression of KIF20A is positively correlated with Ki67 expression, suggesting that KIF20A may participate in the proliferation of gliomas. In addition, we identified that patients with positive KIF20A expression showed more advanced WHO grade. These results prompted us to question the role of KIF20A in gliomas. Therefore, we carried out univariate and multivariate analysis, and our results indicated that KIF20A can act as an independent prognosis factor for the OS of glioma patients; positive KIF20A expression indicates poor clinical outcome. Other prognosis factors include WHO grade as well as surgery procedure.
To further illustrate the role of KIF20A in the regulation of glioma pathogenesis and progression, we explored the expression pattern of KIF20 in different glioma cell lines and performed the siRNA knockdown experiment as well as overexpression experiment. The in vitro results revealed that overexpression of KIF20A can increase the proliferation and invasion capacity of glioma cells, while knockdown of KIF20A can significantly inhibit proliferation and invasion. We also detected that plumbagin, an anticancer drug, can inhibit the expression of KIF20A on both mRNA and protein levels. On the other hand, overexpression of KIF20A in glioma cells can increase the drug resistance to plumbagin, implying that KIF20A could be a valuable chemotherapy target.
To the best of our knowledge, this is the first study demonstrating the expression pattern of KIF20A in gliomas and its clinical significance. Our findings in the present study also revealed part of the mechanisms of its tumor-promoting role in glioma cell lines. In this respect, our study may have considerable clinical implications. However, little is known about the underlying mechanism of KIF20A-regulated plumbagin resistance. It is necessary to carry out more exploratory and validation studies to illuminate the attractive value of KIF20A in the prognostic evaluation and therapeutic strategy of gliomas.
Conclusion
KIF20A can serve as a predictive biomarker for the prognosis of gliomas and may be a potential therapy target.
Acknowledgment
We are grateful to Mary Hannah Swaney from the Pennsylvania State University for helpful writing and editing.
Disclosure
The authors report no conflicts of interest in this work.
References
Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. | ||
Thorne AH, Meisen WH, Russell L, et al. Role of cysteine-rich 61 protein (CCN1) in macrophage-mediated oncolytic herpes simplex virus clearance. Mol Ther. 2014;22(9):1678–1687. | ||
Yang P, Wang Y, Peng X, et al. Management and survival rates in patients with glioma in China (2004–2010): a retrospective study from a single-institution. J Neurooncol. 2013;113(2):259–266. | ||
Kraus JA, Lamszus K, Glesmann N, et al. Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol. 2001;101(4):311–320. | ||
Sarli V, Giannis A. Inhibitors of mitotic kinesins: next-generation antimitotics. ChemMedChem. 2006;1(3):293–298. | ||
Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407(6800):41–47. | ||
Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682–696. | ||
Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. | ||
Fontijn RD, Goud B, Echard A, et al. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol Cell Biol. 2001;21(8):2944–2955. | ||
Neef R, Gruneberg U, Barr FA. Assay and functional properties of Rabkinesin-6/Rab6-KIFL/MKlp2 in cytokinesis. Methods Enzymol. 2005;403:618–628. | ||
Yamashita J, Fukushima S, Jinnin M, et al. Kinesin family member 20A is a novel melanoma-associated antigen. Acta Derm Venereol. 2012;92(6):593–597. | ||
Lu Y, Liu P, Wen W, et al. Cross-species comparison of orthologous gene expression in human bladder cancer and carcinogen-induced rodent models. Am J Transl Res. 2010;3(1):8–27. | ||
Groth-Pedersen L, Aits S, Corcelle-Termeau E, Petersen NH, Nylandsted J, Jaattela M. Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PloS One. 2012;7(10):e45381. | ||
Khongkow P, Gomes AR, Gong C, et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35(8):990–1002. | ||
Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12(4):539–549. | ||
Imai K, Hirata S, Irie A, et al. Identification of HLA-A2-restricted CTL epitopes of a novel tumour-associated antigen, KIF20A, overexpressed in pancreatic cancer. Br J Cancer. 2011;104(2):300–307. | ||
Taniuchi K, Nakagawa H, Nakamura T, et al. Down-regulation of RAB6KIFL/KIF20A, a kinesin involved with membrane trafficking of discs large homologue 5, can attenuate growth of pancreatic cancer cell. Cancer Res. 2005;65(1):105–112. | ||
Stangel D, Erkan M, Buchholz M, et al. Kif20a inhibition reduces migration and invasion of pancreatic cancer cells. J Surg Res. 2015;197(1):91–100. | ||
Osawa R, Tsunoda T, Yoshimura S, et al. Identification of HLA-A24-restricted novel T Cell epitope peptides derived from P-cadherin and kinesin family member 20A. J Biomed Biotechnol. 2012;2012:848042. | ||
Tomita Y, Yuno A, Tsukamoto H, et al. Identification of promiscuous KIF20A long peptides bearing both CD4+ and CD8+ T-cell epitopes: KIF20A-specific CD4+ T-cell immunity in patients with malignant tumor. Clin Cancer Res. 2013;19(16):4508–4520. | ||
Suzuki N, Hazama S, Ueno T, et al. A phase I clinical trial of vaccination with KIF20A-derived peptide in combination with gemcitabine for patients with advanced pancreatic cancer. J Immunother. 2014;37(1):36–42. | ||
Padhye S, Dandawate P, Yusufi M, Ahmad A, Sarkar FH. Perspectives on medicinal properties of plumbagin and its analogs. Med Res Rev. 2012;32(6):1131–1158. | ||
Niu M, Cai W, Liu H, et al. Plumbagin inhibits growth of gliomas in vivo via suppression of FOXM1 expression. J Pharmacol Sci. 2015;128(3):131–136. | ||
van Rijn SJ, Riemers FM, van den Heuvel D, et al. Expression stability of reference genes for quantitative RT-PCR of healthy and diseased pituitary tissue samples varies between humans, mice, and dogs. Mol Neurob. 2014;49(2):893–899. | ||
Yates JW, Chalmer B, McKegney FP. Evaluation of patients with advanced cancer using the Karnofsky performance status. Cancer. 1980;45(8):2220–2224. | ||
Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. | ||
Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New Engl J Med. 2005;352(10):997–1003. | ||
Pu Y, Li S, Zhang C, Bao Z, Yang Z, Sun L. High expression of CXCR3 is an independent prognostic factor in glioblastoma patients that promotes an invasive phenotype. J Neurooncol. 2015;122(1):43–51. | ||
Taniuchi K, Furihata M, Saibara T. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia. 16(12):1082–1093. | ||
Shi C, Huang D, Lu N, et al. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget. 2016;7(18):26206–26219. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.