Back to Journals » Journal of Inflammation Research » Volume 15
Polyploidy Spectrum Correlates with Immunophenotype and Shapes Hepatocellular Carcinoma Recurrence Following Liver Transplantation
Authors Zhang L, Yang Z, Zhang S, Zhou K, Zhang W, Ling S , Sun R, Tang H, Wen X, Feng X, Song P, Xu X, Xie H, Zheng S
Received 22 October 2021
Accepted for publication 16 December 2021
Published 11 January 2022 Volume 2022:15 Pages 217—233
DOI https://doi.org/10.2147/JIR.S345681
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Liang Zhang,1,* Zhentao Yang,1,* Shiyu Zhang,1 Ke Zhou,1 Wu Zhang,2 Sunbin Ling,1 Ruiqi Sun,1 Hong Tang,1 Xue Wen,3 Xiaowen Feng,1 Penghong Song,1 Xiao Xu,1 Haiyang Xie,1 Shusen Zheng1,2
1Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, The First Affiliated Hospital, Zhejiang University School of Medicine, Key Laboratory of the Diagnosis and Treatment of Organ Transplantation, Research Unit of Collaborative Diagnosis and Treatment for Hepatobiliary and Pancreatic Cancer, Chinese Academy of Medical Sciences (2019RU019), Hangzhou, Zhejiang, 310003, People’s Republic of China; 2Department of Hepatobiliary and Pancreatic Surgery, Shulan (Hangzhou) Hospital, Hangzhou, Zhejiang, 310004, People’s Republic of China; 3Department of Pathology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310003, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Haiyang Xie; Shusen Zheng
School of Medicine, Zhejiang University, 79# Qingchun Road, Hangzhou, Zhejiang, 310000, People’s Republic of China
Tel/Fax +86 571 87236570
; +86 571 87236466
Email [email protected]; [email protected]
Purpose: Patients receiving liver transplantation (LT) for hepatocellular carcinoma (HCC) are at high risk of tumor recurrence. Polyploidy is a fascinating characteristic of the liver and correlates with HCC development and progression. This study aims to investigate the association between hepatocyte polyploidy spectrum and HCC recurrence after LT.
Patients and Methods: Thirty-two paired HCC, peritumoral, cirrhotic, and normal liver specimens were employed to examine the hepatocyte polyploidization pattern during liver tumorigenesis. Clinicopathological implications of polyploidy spectrum for LT recipients with HCC were investigated in 205 patients from two transplant centers. Immunofluorescence staining was performed on paraffin-embedded tissue sections to determine the ploidy profiles in situ. Expression levels of CD4, CD8, forkhead box protein 3 (Foxp3) and programmed death-ligand 1 (PD-L1) were measured using immunohistochemistry. An array-based multiplex ELISA system was used for the quantitative measurement of 40 unique inflammatory cytokines.
Results: The fraction of mononuclear polyploidy increased, whereas that of binuclear polyploidy reduced during hepatocarcinogenesis. Recipients with highly mononuclear polyploid HCC (HMP–HCC) had inferior recurrence-free survival and HCC-specific survival than poorly mononuclear polyploid HCC recipients. These two groups differed in abundance of infiltrative CD8+ cytotoxic T cells and FoxP3+ Treg cells, and PD-L1 expression, as well as circulating granulocyte–macrophage colony-stimulating factor, interferon-γ and interleukin-10 levels. HMP–HCC constituted an independent recurrence predictor and could improve the discriminative efficacy of clinical prediction models (Milan criteria, AFP model, and Metroticket 2.0 criteria). A scoring system incorporating the ploidy signature was developed and validated, allowing for an improved risk prediction relative to the RETREAT score and post-MORAL score.
Conclusion: Polyploid spectra are associated with tumor immunophenotype and provide supplementary prognostic information in LT for HCC.
Keywords: polyploidy, immunosurveillance, tumor-infiltrating lymphocytes, cytokines, tumor recurrence
Introduction
Liver transplantation (LT) is a definitive treatment option for selected patients with hepatocellular carcinoma (HCC), as it simultaneously removes intrahepatic tumors and the underlying carcinogenic liver background.1 Nevertheless, tumor recurrence after LT remains a huge clinical challenge, indicating not only the potential loss of a scare liver graft but also dismal prognosis for the recipient being in an immunocompromised state.1–3 At present, there are still lacks of guidance regarding tailored posttransplant management of this unique population.3 In particular, risk stratification-based surveillance strategies and recommendations regarding recurrence prevention are highly desirable.
Over the past few decades, significant efforts have been made to design scoring systems and individually evaluate recipients’ risk of tumor recurrence.4–7 Unlike traditional rigid binary selection criteria, simple risk scores could provide quantifiable estimation of tumor recurrence risk for a given recipient and potentially assist in the planification of tailored posttransplant management protocols. Several of these risk scores, including the RETREAT score and MORAL score, have also been externally validated.8,9 However, despite the flexibility and robustness, variables employed in the scoring systems to model the recipients’ recurrence risk remain limited, particularly in terms of tumor biology measures. Thus, identifying novel and reliable surrogates for HCC tumor aggressiveness would capture additional information, and integrating it for recurrence risk estimation may help to further improve the prognostic efficacy, better guide individualized risk-based management strategies, and optimize transplant outcomes.
Polyploidy, or whole-genome doubling, is a characteristic feature of the liver, wherein cells acquire additional sets of chromosomes.10 In mammals, whole-organism polyploidization typically causes fetal lethality, although several tissues including the liver parenchyma develop a certain degree of polyploidy.11 Hepatocyte polyploidization could be either characterized by the generation of two or more nuclei per cell, defining cellular polyploidy, or completed within a single nucleus, defining nuclear polyploidy. As many as 30% of hepatocytes in the adult human liver are polyploid, and this ratio even increases to nearly 90% in rodents.12 Ploidy profiles of the liver are not set in stone, while it would display dramatic changes both during physiological processes and in pathological conditions, including the liver development, senescence, tissue regeneration, genotoxic stress, and metabolic overload.10,12
Hepatocyte ploidy spectra alterations correlated with the onset and progression of non-alcoholic fatty liver disease, Wilson’s disease, chronic viral hepatitis and its complications.13–15 Experimental evidence has also been accumulating identifying unscheduled polyploidization as a crucial contributor to cancer genomic instability and a key gatekeeper during the development of HCC.16 During the examination of human liver specimen, a recent study suggested the use of polyploidy spectrum as a promising recurrence predictor for clinical HCC patients receiving liver resection, advocating it as a novel hallmark in HCC classification.17 However, until now, the prognostic value of liver ploidy profiles in the setting of LT for HCC has not been explored.
One determinant factor being increasingly recognized to accelerate the acquisition of a more aggressive phenotype of HCC is immune evasion, which protects tumor cells from immune system recognition and destruction. Tumor-infiltrating lymphocytes (TILs), the primary immune component infiltrating tumor tissues, represent the manifestation of the host anti-tumor response and have been shown to be closely associated with HCC recurrence.18–20 During the activation of immune responses, inflammatory cytokines, a broad category of small soluble proteins (usually <30 kDa), are released and play a crucial role in mediating intercellular communications, antigen priming, and cytolytic activities of effector immune cells.21 Adjuvant cytokine therapies also emerge as an effective strategy to enhance anti-tumor immunity.22 Although polyploidization has been considered a critical step in tumor development and progression, the association between ploidy profiles and tumor immune surveillance in HCC patients remains unclear.
In this study, we assessed the feasibility and efficacy of utilizing polyploidy profiles as a novel metric to stratify recurrence risk for LT recipients with HCC, and suggested the relationship between modification of ploidy status and immune phenotype in these patients.
Materials and Methods
Study Population
Three cohorts of participants were enrolled in this study, and the corresponding formalin-fixed and paraffin-embedded liver tissues were collected. Thirty-two pairs of HCC, cirrhosis, and health controls composed of the observation set to characterize hepatic ploidy alterations during liver tumorigenesis. Both tumoral and peritumoral liver tissues were obtained for HCC. Demographic characteristics, including gender and age, were matched among the groups. The tissue samples were collected from the biobank at First Affiliated Hospital, School of Medicine, Zhejiang University.
The derivation set was designed to evaluate the correlation between hepatocyte polyploidy spectrum and tumor characteristics of LT recipients with HCC, and to develop a prognostic scoring model for recurrence prediction. This set composed 83 adult patients (>18 years) undergoing LT for HCC from February 2015 to November 2017 at the First Affiliated Hospital, School of Medicine, Zhejiang University, and pretransplant plasma samples were collected. Excluded were multiple-organ transplantation, split LT, re-transplantation, missing HCC specimens or follow-up data, and no viable tumor in explant pathology. For the purpose of external validation of the prediction model, another 122 transplant recipients with HCC at the Shulan (Hangzhou) Hospital from April 2016 to January 2019 constituted the validation set, following the application of the same inclusion and exclusion criteria as in the derivation set.
Data Collection
Clinical baseline details were gathered including recipient age and gender, Child-Pugh score, model for end-stage liver disease score, underlying hepatic cirrhosis and etiological risk factors. Tumor characteristics, including the tumor size, number, histopathological differentiation according to Edmondson-Steiner criteria (grading I–II or III–IV), and microvascular invasion (MVI, with or without) were determined by the explant pathological findings. The cut-offs of maximum tumor diameter (≤5 cm/>5 cm), tumor number (≤3/>3), serum AFP level (≤400 ng/mL/>400 ng/mL), neutrophil lymphocyte ratio (<5/≥5), platelet lymphocyte ratio (<150/≥150), and lymphocyte monocyte ratio (<2.75/≥2.75) were considered in accordance with previous publications.1,23–25
The selection criteria and posttransplant models, including the Milan criteria, the Metroticket 2.0 criteria, the AFP model, the post-MORAL score (Model Of Recurrence After Liver transplant), and the RETREAT score (Risk Estimation of Tumor Recurrence After Transplant) were all determined based on the pathological examination of explants at the time of LT. They were defined according to the previous formulas.1,4,6,26,27
Perioperative Management and Follow-Up
The perioperative management protocol and posttransplant surveillance strategy are similar at the two centers and have been described previously.28,29 In brief, all recipients received basiliximab induction therapy (20 mg, within two hours before LT and on postoperative day four) and intraoperative methylprednisolone (500 or 1000 mg). The posttransplant immunosuppression regime was steroid-free and consisted of a combination of tacrolimus and mycophenolate mofetil. The surveillance of recurrence included the measurement of serum AFP, ultrasonography, and computed tomography (every 3 months for the first two years post-transplant and biannually thereafter). Tumor recurrence was diagnosed by positive radiological findings and elevation of serum AFP.
Immunofluorescence and Qualification of Mononuclear and Binuclear Ploidy
The 5-μm tissue slides were deparaffinized and rehydrated, and a 3% H2O2 solution was used for endogenous peroxidase inactivation. Antigen retrieval was heat-induced in the citrate buffer solution (pH 6.0). Then, the slides were blocked with normal fetal bovine serum (37°C), and subsequently incubated with β-catenin (dilution 1:200; BD Biosciences, Cat#610154) overnight at 4°C. The secondary antibody was the anti-mouse IgG antibodies conjugated with Alexa Fluor 488 (1:200; Cell signaling Technology, Cat#4408S), and 4,6-diamidino-2-phenylindole (DAPI; Zhong Shan Golden Bridge Biological Technology Inc., Beijing, China) was used to counterstain nuclei.
Hepatocyte ploidy profiles were determined in situ on immunofluorescence-stained liver tissue sections, according to the previously described method.13,15,17,30,31 Immunofluorescence images were captured using an automated fluorescence microscope (BX63, Olympus, Tokyo, Japan). Image J (NIH, USA) was used for quantitative image analysis. The images were adjusted in 8 bits and a nuclear circularity ≥0.7 was applied to exclude non-epithelial cell populations. Binuclear polyploidy was recognized by the number of nuclei per hepatocyte, mononuclear polyploidy was characterized on the ground of the area of the nuclei, and the number of evaluated tumor cells was reported as respective denominators. Hepatocytes with the nuclear areas of 200–750 pixel2, 800–1350 pixel2 and 1400–4000 pixel2 were defined as diploid (2n), mononuclear tetraploid (4n) and octoploid (≥8n), respectively, according to the distribution histogram of nuclear area (Supplementary Figure 1); for the vast majority (>90%) of examined cases, the histogram consistently revealed a clear trimodal distribution. Nuclear area <200 pixel2 or >4000 pixel2 were excluded, due to neither non-hepatocytes population nor incorrect DAPI segmentation. The mononuclear ≥8n fraction in most cases was too small (only accounting for <5% of the total hepatocyte population), and therefore we, exploiting from previous experiences,15,17 amalgamated the mononuclear 4n and ≥8n fractions into the fraction of mononuclear polyploidy. For cellular ploidy analysis, 10–15 random high-power fields were selected, and about 2000 cells were counted from each slide. For nuclear ploidy analysis, more than 5000 cells were counted for each patient.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described.32,33 The sections were stained for CD4 (1:500; Abcam, ab133616), CD8 (1:50; Abcam, ab237709), forkhead box P3 (FoxP3; 1:200; Abcam, ab20034) and programmed cell death-ligand 1 (PD-L1; 1:200; Abcam, ab205921). The density of CD4+ Th cells, CD8+ cytotoxic T cells, and FoxP3+ Treg cells (number/mm2) was determined as the average number of immunoreactive cells under the 200× magnification fields. At least eight randomly chosen and non-overlapping fields were required for the evaluation, with avoidance of peripheral regions on each slide. PD-L1 expression was semi-quantitatively evaluated considering both tumor and immune stroma. Specifically, positive tumoral PD-L1 expression was considered when more than 1% of neoplastic cells displayed membranous staining.34 For PD-L1 expression in the immune stroma, any unequivocal expression of PD-L1 (>0%) on stromal immune cells was considered to be positive.35 Examples of PD-L1 IHC evaluation are depicted in Supplementary Figure 1.
Inflammatory Cytokine Analysis
Peripheral blood samples were collected from recipients 1–2 days prior to transplant, and then centrifuged for 10 minutes at 3000 rpm at 4°C for serum and plasma separation. The plasma was harvested, divided into 1 mL aliquots, and stored at −80°C until analysis. A commercially available array-based multiplex sandwich ELISA kit (Human Inflammation Array Q3; QAH-INF-3, RayBiotech, Inc. Norcross, GA, USA) was used to measure the concentrations of 40 inflammatory cytokines according to the manufacturer’s protocol; these cytokines included B-lymphocyte chemoattractant, eotaxin, eotaxin-2, granulocyte colony-stimulating factor, GM-CSF, I-309, intercellular adhesion molecule-1, IFN-γ, IL-1α, IL-1β, IL1 receptor antagonist, IL-2, IL-4, IL-5, IL-6 and its soluble receptor, IL-7, IL-8, IL-10, IL-11, IL-12p40, IL-12p70, IL-13, IL-15, IL-16, IL-17, monocyte chemotactic protein −1, macrophage colony-stimulating factor, monokine induced by gamma interferon, macrophage inflammatory protein (MIP)-1α, MIP-1β, MIP-1δ, platelet-derived growth factor-BB, regulated on activation normal T cell expressed and secreted, tissue inhibitor of metalloproteinase (TIMP)-1, TIMP-2, tumor necrosis factor (TNF)-α, TNF-β, and soluble receptors TNF-sRI and TNF-sRII.
Statistical Analysis
The normality was determined using the Shapiro–Wilk test. Normally distributed variables are presented as the mean ± standard deviation and compared using the Student’s t-test. Skewed variables are reported as the median with IQR and compared using the Wilcoxon rank sum test or Dunn’s multiple comparisons test, as appropriate. Bivariate correlation analysis was conducted using Spearman’s rank-order correlation. Categorical variables are described as numbers (percentages), and Pearson’s chi-square test or Fisher’s exact test was used for comparisons. Recurrence-free survival was set as the primary outcome measure and defined as the time elapsed from LT to either tumor recurrence or censoring at the last follow-up. A competing-risk analysis was used to evaluate the HCC-specific survival, defined as the interval from LT to either mortality with a documented HCC recurrence or last follow-up.26 Survival curves were computed using the Kaplan–Meier method and compared by the Log rank test. In the validation set, due to the relatively limited follow-up length and no recipient had been followed up for more than five years, the estimated five-year survival rate was not computed by the Kaplan–Meier plot.
Univariate and subsequent backward stepwise multivariate Cox regression analyses were performed to identify independent prognostic factors. A model predictive of post-LT recurrence was developed using the independent predictors weighted by rounding the coefficients in the multivariable model to the nearest integer. The area under the receiver operating characteristic (ROC) curve was calculated to assess the predictive efficacy of mathematical models and compared using the Hanley and McNeil method. Statistical analysis was carried out using the R-project (version 3.6.1, https://www.r‐project.org/) and MedCalc (MedCalc Software; Mariakerke, Belgium). All p-values were two-tailed, and p <0.05 was considered statistically significant.
Results
Participant Characteristics
This study included three independent cohorts of patients (Table 1). The mean age of patients in the observation, derivation and validation sets was 51.6 ± 10.6, 52.3 ± 9.3, and 49.9 ± 11.2 years, and 96.9%, 96.4% and 89.3% are male, respectively. 96.2% of patients had hepatitis B virus infection. The median follow-up for the LT recipients with HCC in the derivation set was 23.9 months (interquartile range [IQR], 12.7–39.4) and in the validation set was 18.9 months (IQR, 8.4–29.4). In terms of recurrence-free survival and HCC-specific survival, HCC recipients of both two transplant cohorts were well stratified by the Milan criteria, AFP model, and Metroticket 2.0 criteria (all p<0.05; Supplementary Figures 2 and 3).
 |
Table 1 Demographics and Tumor Characteristics of the Study Population |
Alterations of Hepatocyte Polyploidization During Hepatocarcinogenesis
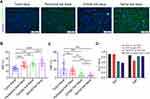
To evaluate the hepatocyte ploidisation pattern during hepatocarcinogenesis, we analyzed 32 paired HCC, peritumoral, cirrhotic, and normal liver tissues from the observation set (Figure 1A). The results showed that during liver tumorigenesis, the fraction of binuclear polyploidy (FBP) was drastically decreased (Figure 1B), whereas the fraction of mononuclear polyploidy (FMP) was increased (Figure 1C), which is in agreement with previous observations.17 We also examined the alterations of ploidy content of steatotic livers. Of the normal livers, 6 were steatotic. Compared to non-steatotic livers, steatotic livers had significantly reduced FBP (12.7% vs 9.1%, p=0.01), while there was no significant difference in FMP (5.8% vs 6.3%, p=0.74).
To further evaluate whether polyploidy spectrum could act as a classification signal, the ROC curve was constructed, and the area under the curve (AUC) was determined (Figure 1D). FMP could distinguish HCC and peritumoral tissues from non-HCC tissues (AUC = 0.868 and 0.827, respectively), while the diagnosis efficacy was weak between HCC and peritumoral tissues (AUC = 0.658). FBP was able to distinguish HCC and non-HCC tissues from peritumoral tissues (AUC = 0.916 and 0.905, respectively), but not peritumoral tissues from non-HCC tissues (AUC = 0.636). Interestingly, FMP could further effectively predict the presence of MVI in HCC tissues (AUC = 0.826), whereas FBP failed (AUC = 0.575), indicating that FMP may be more sensitive to advanced HCC.
FMP Predicts Post-LT Tumor Recurrence
The association between polyploidy spectrum and HCC recurrence after LT was investigated. Of 83 HCC recipients in the derivation set, the median FBP and FMP were 7.7% (6.5%–9.2%) and 17.8% (8.8%–32.1%), respectively. Univariate Cox analysis showed that FBP had no statistical correlation with the risk of recurrence as a continuous variable (OR = 0.92, 95% CI, 0.804–1.052, p=0.23). In contrast to FBP, each percent increase in FMP as a continuous parameter was significantly associated with an OR of 1.035 for increased risk of tumor recurrence (95% CI, 1.015–1.056, p=0.001). The distribution of FBP was similar between LT recipients with and without recurrence (7.1% vs 8.3%, p=0.15), whereas LT recipients who experienced HCC recurrence had significantly higher FMP relative to recipients who did not (25.1% vs 10.5%, p<0.001). Furthermore, the AUC for FBP in forecasting the possibility of posttransplant recurrence was only 0.57, while for FMP was 0.742 (Hanley and McNeil test, p=0.04).
The ROC curve selected an optimum threshold value of 18% for FMP to predict recurrence, and accordingly, two subtypes of HCC were defined: highly mononuclear polyploid HCC (HMP–HCC, FMP > 18%) and poorly mononuclear polyploid HCC (PMP–HCC, FMP ≤ 18%). Of 44 HMP–HCC patients, 31 (70.5%) developed HCC recurrence, compared to 11 of 39 (28.2%) PMP–HCC patients (p<0.001). Patients with PMP–HCC displayed significantly better recurrence-free survival and HCC-specific survival than those with HMP–HCC (p<0.001 and p=0.02, respectively; Figure 2A and Supplementary Figure 4A).
In univariate Cox regression analysis, maximum nodule diameter >5 cm, nodule number >3, histopathologic grading III–IV, MVI, serum AFP >400 ng/mL, HMP–HCC, neutrophil lymphocyte ratio ≥5, and platelet lymphocyte ratio ≥150 were associated with increased risks of HCC recurrence (Table 2). In the multivariate model, maximum nodule diameter >5 cm, nodule number >3, HMP–HCC, and AFP > 400 ng/mL constituted four independent risk factors.
 |
Table 2 Cox Analyses of Variables Related to the Recurrence-Free Survival |
FMP Provides Supplementary Discriminative Information
The clinicopathological parameters were compared between patients with HMP–HCC and PMP–HCC (Table 3). Compared to patients with PMP–HCC, those with HMP–HCC had numerically higher proportion of nodule number >3, histopathologic grading III–IV, and MVI, as well as AFP > 400 ng/mL, suggesting less favorable tumor biology in HMP–HCC patients.
 |
Table 3 Comparisons of Clinicopathological Features Between Patients with PMP–HCC and HMP–HCC |
Among patients within the Milan criteria, those with HMP–HCC and PMP–HCC had similar recurrence-free survival and HCC-specific survival (p=0.62 and 0.27, respectively). Whereas, for patients outside the Milan criteria, those with PMP–HCC displayed significantly better recurrence-free survival and HCC-specific survival than HMP–HCC (p=0.001 and 0.04, respectively; Figure 2B and Supplementary Figure 4B). Furthermore, patients who exceeded the Milan criteria but had PMP–HCC exhibited comparable recurrence-free survival and HCC-specific survival as compared to those fulfilling the Milan criteria (p=0.61 and 0.30, respectively). In the subgroup beyond the Milan criteria, HMP–HCC patients had a significantly increased proportion of nodule number >3 relative to PMP–HCC patients (64.9% vs 28.6%, p=0.01); HMP–HCC also tended to have a higher frequency of MVI, although this difference failed to reach significance (59.5% vs 33.3%, p=0.06).
PMP–HCC patients with AFP score >2 had comparable recurrence-free survival and HCC-specific survival relative to patients who had AFP score ≤2 (p=0.61 and 0.94, respectively), but better than those HMP-HCC patients with AFP score >2 (p=0.02 and 0.05, respectively; Figure 2C and Supplementary Figure 4C). Likewise, the recurrence-free survival and HCC-specific survival of patients who exceeded the Metroticket 2.0 criteria but had PMP–HCC were also similar to those patients fulfilling the Metroticket 2.0 criteria (p=0.23 and 0.14, respectively; Figure 2D and Supplementary Figure 4D).
HMP–HCC Correlates with an Immunosuppressive Phenotype
To determine whether amplification of nuclear ploidy was associated with the tumor immune microenvironment, we measured the densities of intratumoral CD4+ helper T (Th) cells, FoxP3+ T regulatory (Treg) cells, and CD8+ cytotoxic T cells, and PD-L1 expression.
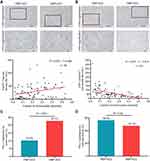
Compared to PMP–HCC, HMP–HCC had significantly higher density of Foxp3+ Treg cells but lower density of CD8+ cytotoxic T cells (Figure 3A and B and Supplementary Figure 5A and B). In a continuous manner, FMP was positively correlated with the density of FoxP3+ Treg cells (r=0.365, p=0.001) and negatively correlated with that of CD8+ cytotoxic T cells (r=−0.614, p<0.001). Whereas, the density of CD4+ Th cells were similar between PMP–HCC and HMP–HCC; the density of CD4+ Th cells showed no significant correlation with FMP as a continuous variable (r=0.05, p=0.65; Supplementary Figure 5C and D). Replacing the density of CD8+ T cells with the CD8:CD4 ratio generated similar correlation with FMP (r=−0.597, p<0.001), as well that of FoxP3+ T cells with the Foxp3:CD4 ratio (r=−0.336, p<0.001).
HMP–HCC also had a higher frequency of PD-L1 expression by the tumor cells than PMP–HCC (Figure 3C), whereas PD-L1 expression in the immune stroma did not differ between the two groups (Figure 3D).
As a critical component of the immune system, cytokines mediate intercellular communications and help shape anti-tumor immune responses. To investigate the relationship between mononuclear polyploidy and circulating cytokine levels, we measured the plasma concentrations of a panel of 40 cytokines (described in Methods). As shown in Table 4, compared to patients with PMP–HCC, those with HMP–HCC had significantly higher levels of interleukin (IL)-10 (p=0.03) while lower interferon (IFN)-γ (p=0.02) and granulocyte–macrophage colony-stimulating factor (GM-CSF) (p=0.01). HMP–HCC also tended to be associated with increased IL-6 levels and reduced IL-2 levels than PMP–HCC, while these differences were not significant (p=0.06 and 0.09, respectively).
 |
Table 4 Comparisons of Circulating Inflammatory Cytokine Concentrations Between Patients with HMP–HCC and PMP–HCC |
Risk Assessment Model Incorporating the Ploidy Signature
The predictive value of ploidy profiles of tumor recurrence were further evaluated by constructing a simplified risk assessment model incorporating the ploidy signature (RAMPS), and recipients were re-classified into four groups for risk stratifications. Nineteen patients in the absence of any risk factors were allocated with a RAMPS score of 0 and were included in the low-risk group, 16 patients presenting with only one factor had a score of 4–6 and were in the medium risk group, 24 with two factors had a score of 9–11 and in the high-risk group, and the remaining 24 with three or more risk factors were scored 14–20 and included in the very high-risk group.
As depicted in Figure 4A, the low-risk group achieved the best prognosis, while the very high-risk group displayed the worst. Differences between recurrence-free survival in each of the groups were all significant (p<0.05), except the comparison between the low and medium risk groups (p=0.33).
The RAMPS score was then tested in the validation set. Of this set, the median FMP was 10.8% (5.5%–21.3%), and 37 recipients had HMP–HCC. A significantly increased risk of HCC recurrence was noted in HMP–HCC patients relative to PMP–HCC patients (p<0.001). Comparisons of recurrence risk between HMP–HCC and PMP–HCC in subgroups stratified by the Milan criteria, the Metroticket 2.0 criteria, and the AFP model yielded very similar results to that in the derivation set (Supplementary Figures 6 and 7). The RAMPS score again allowed clear stratification of recurrence-free survival among the four groups in the validation set (Figure 4B).
The prognostic performance of RAMPS score was further compared to that of two well-established posttransplant models—the RETREAT score and post-MORAL score. In the derivation set, the RAMPS score achieved the highest AUC values for the predictions of 3- and 5-year recurrence when compared to the RETREAT score and post-MORAL score (Figure 5A). The parallel evaluation on validation set revealed that the RAMPS score maintained the highest AUC values than the other two prognostic models for prediction of tumor recurrence (Figure 5B).
Discussion
Polyploid hepatocytes have been documented for more than a century. However, the clinicopathological relevance of hepatocyte polyploidy spectrum in patients with HCC remains to be fully explored. This study demonstrates that hepatocyte polyploidy spectrum can serve as a useful biological metric to estimate posttransplant tumor recurrence risk. Our study also provides evidence linking genome instability and tumor immune evasion in HCC patients, and develops a new tool for prognostic stratification after LT for HCC.
Genomic instability represents a hallmark of most cancers.36 As one of the most frequent genomic events, polyploidization of cancer cells favors aberrant mitoses leading to blocked or asymmetric cell division, generates genetic diversity, and accelerates the cancer genome evolution.10,11,16,37 Triggered by continuous replication stress, polyploid cancer cells present with ongoing chromosomal instability and display enhanced aggressiveness.38 Indeed, most solid tumors exhibit polyploid karyotypes.37,39 A previous pan-cancer analysis involving 4934 tissue specimens across 11 cancer types indicated that whole-genome doubling occurred in 37% of cancers.39 Furthermore, polyploidization has been shown to correlate with inferior prognosis in many types of malignancies, including colorectal cancer, pancreatic cancer, and breast cancer.37 Nevertheless, data on HCC patients were not available in these studies. An exquisite series analysis of 75 surgically resected HCC by Bou-Nader et al reported that amplification of nuclear ploidy was related to increased proliferative activity, TP53 mutation, and poor histopathological differentiation, as well as shortened disease-free survival; of interest, they also found that polyploid hepatocytes do not display a specific zonal distribution in hepatic lobules.17 Likewise, our study also observed a correlation between the amplified nuclear ploidy and unfavorable HCC clinicopathological characteristics. Compared to patients with PMP–HCC, those with HMP–HCC were more likely to have multifocal nodules, poor pathological differentiation, MVI, high serum AFP level, and inferior oncological outcomes. Taken together, our data parallel previous findings and support the notion that whole-genome duplication is closely associated with the prognosis of patients with advanced cancers.
An interesting and novel finding in this study is that we noted a close correlation between polyploidy spectrum and immune response signature in HCC patients. The mononuclear polyploid fraction was positively correlated with the densities of CD8+ cytotoxic T cells while negatively with that of FoxP3+ Treg cells. The amplification of mononuclear polyploidy also displayed the upregulation of tumoral PD-L1 expression. Moreover, significantly increased secretion of immunosuppressive cytokines (IL-10) while reducing anti-tumoral cytokines (IFN-γ and GM-CSF) further provided evidence of an immunosuppressive phenotype in HCC patients with amplified mononuclear polyploidy. Intratumoral FoxP3+ Treg cells density, PD-L1 expression and circulating IL-10 have shown to inversely correlate with clinical outcomes of HCC patients, while increased CD8+ T lymphocytes density and circulating IFN-γ were reported as protective factors.20,34,40 In line with our observations, Senovilla et al analyzed ploidy spectra in clinical samples of 60 patients with breast cancer, and noted a negative correlation between polyploidy profiles and the ratio of CD8+ cytotoxic T cells to Foxp3+ Treg cells.41 Studies on rodent models also documented that hyperploid cancer cells would conserve their ploidy and readily proliferate when growing in immunodeficient mice, whereas tumors arising in immunocompetent hosts showed decreased ploidy and retarded growth.41,42 Mechanically, polyploidy cancer cells displayed an exacerbated endoplasmic reticulum stress, which induced calreticulin expression on the cell surface and eventually triggered the immune elimination of hyperploid cells. The composition of TILs and immune effectors, including CD8+ and FoxP3+ T lymphocytes, immune checkpoints, and interferon-γ, have been implied in the immunosurveillance of cancer cell ploidy, which coincides with tight associations observed in our study.43 Therefore, in addition to cell-intrinsic monitoring patterns, an extrinsic immune mechanism also plays an important role in maintaining the genome integrity.43
Modification of hepatocyte ploidy content was implied during HCC development. So far, only one clinical study had systematically analyzed the hepatocyte ploidisation pattern including the cellular and nuclear ploidy during human hepatocarcinogenesis, and our values showed similar proportions of the fraction of mono- and bi-nuclear polyploid cells. Through the examination of tumoral and peritumoral tissues from HCC patients and normal liver tissues, Bou-Nader et al suggested that liver tumorigenesis was associated with dramatically decreased cellular ploidy while increasing nuclear ploidy.17 The present study confirmed these findings and further added data on hepatocyte polyploidy content in cirrhotic stage. Consistent with these observations in clinical samples, recent experimental assays in rodent models revealed that hyperpolyploidization of hepatocyte constituted an early and critical step in malignant transformation.16 Interestingly, in a panel of rodent models of chronic liver injury, decreased cellular ploidy, while increased nuclear ploidy was also observed, which suggests that polyploidization may precede hepatocarcinogenesis.44 Likewise, in a chemical carcinogen-induced HCC rodent model, Sladky et al also found that cellular polyploid was significantly reduced in tumor tissues compared with normal liver tissues.45 Whereas, in a subsequent clinical cohort of HCC patients in the same work, the authors used the number of cells per field (cell density) as a surrogate marker of ploidy, which did not distinguish cellular ploidy from nuclear ploidy and would include non-epithelial cells into considerations. In addition, we also evaluated the discriminative performance of the polyploidy spectrum as a classification signal. Of interest, we found a weak discriminative efficacy for FMP to distinguish HCC from peritumoral tissues but a good performance to predict the presence of MVI in HCC tissues, which suggested that advanced HCC is likely a major contributor to the observed changes in FMP during hepatocarcinogenesis. In contrast, FBP might be more sensitive to relatively early-stage HCC, as which effectively distinguished HCC from non-HCC tissues and peritumoral tissues but whose diagnostic power was weak for the prediction of MVI.
Despite these evidences documenting ploidy content modifications in HCC tissues and clinical observations, including the present study highlighting the amplified ploidy content as an unfavorable prognostic indicator in patients with HCC, this is certainly not to conclude that polyploidization promotes the development of HCC due to the nature of observational studies. Instead, several experimental assays have indicated a tumor-suppressive role of the polyploid state in the development of HCC. In rodent HCC models, manipulation of ploidy content by interfering cytokinesis-related genes could effectively inhibit the tumor growth, while it did not significantly compromise the liver regenerative and proliferative activities as well as liver functions, rendering it as a novel therapeutic strategy.46,47 The mechanism by which polyploid HCC cells generate and can act as a negative prognostic factor, while rodents with increased polyploidy hepatocyte could suppress tumorigenesis, still remains a mystery and warrants further research.48
Alterations of ploidy spectra have also been reported in nonalcoholic fatty liver disease, a rapidly rising risk factor contributing to HCC development in Western countries.49 Gentric et al suggested that non-alcoholic steatohepatitis was associated with dramatically increased FMP and reduced FBP.13 Likewise, we observed that steatotic livers exhibited lower FBP than non-steatotic livers. The FMP, however, did not differ by the presence of steatosis. One possible explanation to this discrepancy was due to the characteristics of the participants. In the prior clinical cohort, quantification of nuclear ploidy for the fatty liver was carried out in 24 patients with severe metabolic syndrome (of whom 16 had concomitant HCC), indicating a very advanced stage of disease.13 Meanwhile, in the present study, we controlled the presence of steatosis as the only variable in the histologically normal livers. Alternatively, our results may suggest that the modification of cellular ploidy precedes that of nuclear ploidy during the natural course of non-alcoholic fatty liver disease. Nevertheless, given the limited number of steatotic cases, further investigations with larger sample sizes and varying stages of steatosis are warranted to confirm these findings.
Polyploidy spectrum could refine the prognostic performance of clinical prediction models (Milan criteria, Metroticket 2.0 criteria and AFP model). Moreover, multivariate analysis indicated that HMP–HCC conferred up to a two-fold increased risk of recurrence. Tumor number, diameter, and AFP were identified as other three independent risk factors, and they were identical to variables composing the vast majority of previous prognostic indexes.6,26,27,50 To this end, the RAMPS score was developed and externally validated; by accounting for the tumor ploidy signature, morphological burden and serum AFP level, this scoring system could provide a hierarchy of possibilities of tumor recurrence, categorize recipients into four different risk groups, and attain higher predictive performance than the RETREAT score and post-MORAL score. The RAMPS score seems to be a promising tool not only to inform LT recipients more precisely about the expected incidence of HCC recurrence and tailor their immunosuppression and frequency of surveillance imaging accordingly but also to help identify those recipients at high risk for tumor recurrence for the enrollment into future clinical trial of posttransplant adjuvant therapies.
There are several limitations that should be acknowledged. First, this investigation was based on retrospective cohorts, and potential selection bias could not be eliminated, although this is, to our knowledge, the largest series examining the clinicopathological relevance of polyploid spectrum in HCC patients. The majority of the study population had hepatitis B virus infection. Further investigations with various risk factors of HCC are essential to verify our findings. Second, due to lack of data, we failed to assess the effect of response to bridging locoregional therapy on posttransplant outcomes, which is also an identifier of tumor biology and an important factor determining transplant eligibility.51 However, this would probably not have added much more information, as our study is not an intention-to-treat analysis. Third, the transplant recipients received the same immunosuppressant protocol (without sirolimus), which hampered the comparison of the oncological effects between sirolimus and calcineurin inhibitors, as well the potential implications of ploidy signature for the management of immunosuppression. We are in the process of enrolling another transplant cohort to further investigate whether polyploid spectrum is useful in determining the appropriate immunosuppression regimen after LT for HCC.
Conclusion
In summary, our findings demonstrated that hepatocyte polyploid spectrum correlated with tumor immunosurveillance and delivered supplementary prognostic information for recipients who underwent LT for HCC. We developed and validated a scoring system, which is a potentially important tool for an optimal estimation of the recipients’ tumor recurrence risk and facilitation of an individualized posttransplant management strategy.
Abbreviations
AFP, α-fetoprotein; AUC, area under the curve; CI, confidence interval; FBP, fraction of binuclear polyploidy; FMP, fraction of mononuclear polyploidy; GM-CSF, granulocyte–macrophage colony-stimulating factor; HCC, hepatocellular carcinoma; HMP–HCC, highly mononuclear polyploid hepatocellular carcinoma; IL, interleukin; IFN, interferon; LT, liver transplantation; MIP, macrophage inflammatory protein; MVI, microvascular invasion; OR, odds ratio; PD-L1, programmed death-ligand 1; PMP–HCC, poorly mononuclear polyploid hepatocellular carcinoma; RAMPS, risk assessment model incorporating the ploidy signature; TIMP, tissue inhibitor of metalloproteinase; TNF, tumor necrosis factor; ROC, receiver operating characteristic; TILs, tumor-infiltrating lymphocytes.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author (Shusen Zheng) upon reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (2020-763) and the Shulan (Hangzhou) hospital (2020-074). Written informed consent was obtained from all donors and recipients prior to transplantation. Each organ donation or transplant was approved by the Institutional Review Board at the corresponding centers, under the guidelines of the Ethics Committee of the hospitals, the current regulation of the Chinese Government, and the 1975 Declaration of Helsinki. All liver grafts came from deceased donors. No organs were harvested from executed prisoners.
Consent for Publication
Informed consent was obtained from all individual participants included in the study.
Acknowledgments
The authors thank Leiming Cheng, Hai Cheng, Guoqiang Cao, and Biao Li for their assistance in the management of tissue samples.
Author Contributions
L. Zhang, Z. Yang, H. Xie, and S. Zheng designed the study, analyzed the data, and wrote the article. S. Zhang and R. Sun performed the immunohistochemistry staining. L. Zhang, Z. Yang, and S. Zhang performed the immunofluorescence staining and cytokine analysis. H. Tang, X. Wen, and X. Feng reviewed the histological slides. L. Zhang, K. Zhou, W. Zhang, and S. Ling collected the data. X. Xu, P. Song, and H. Xie contributed to the data interpretation and critical appraisal. H. Xie and S. Zheng revised the article and approved the final version. This article has been approved by all authors listed for publication. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This investigation was supported by the National Science and Technology Major Project (2017ZX10203205), the National Natural Science Foundation of China (81721091), and the Project from the Health Commission of Zhejiang Province (JBZX-202004).
Disclosure
The authors declare no conflicts of interest in this study.
References
1. Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334(11):693–699.
2. Mehta N, Bhangui P, Yao FY, et al. Liver transplantation for hepatocellular carcinoma. working group report from the ILTS Transplant Oncology Consensus Conference. Transplantation. 2020;104(6):1136–1142.
3. Verna EC, Patel YA, Aggarwal A, et al. Liver transplantation for hepatocellular carcinoma: management after the transplant. Am J Transplant. 2020;20(2):333–347.
4. Mehta N, Heimbach J, Harnois DM, et al. Validation of a Risk Estimation of Tumor Recurrence After Transplant (RETREAT) score for hepatocellular carcinoma recurrence after liver transplant. JAMA Oncol. 2017;3(4):493–500.
5. Lai Q, Nicolini D, Inostroza Nunez M, et al. A novel prognostic index in patients with hepatocellular cancer waiting for liver transplantation: time-Radiological-response-Alpha-fetoprotein-INflammation (TRAIN) score. Ann Surg. 2016;264(5):787–796.
6. Halazun KJ, Najjar M, Abdelmessih RM, et al. Recurrence after liver transplantation for hepatocellular carcinoma: a new MORAL to the story. Ann Surg. 2017;265(3):557–564.
7. Sasaki K, Firl DJ, Hashimoto K, et al. Development and validation of the HALT-HCC score to predict mortality in liver transplant recipients with hepatocellular carcinoma: a retrospective cohort analysis. Lancet Gastroenterol Hepatol. 2017;2(8):595–603.
8. Mehta N, Dodge JL, Roberts JP, Yao FY. Validation of the prognostic power of the RETREAT score for hepatocellular carcinoma recurrence using the UNOS database. Am J Transplant. 2018;18(5):1206–1213.
9. Chang Y, Cho Y, Lee JH, et al. Comparison of models for tumor recurrence after liver transplantation for the patients with hepatocellular carcinoma: a multicenter long-term follow-up study. Cancers (Basel). 2019;11:9.
10. Donne R, Saroul-Ainama M, Cordier P, Celton-Morizur S, Desdouets C. Polyploidy in liver development, homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17(7):391–405.
11. Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610.
12. Wang MJ, Chen F, Lau JTY, Hu YP. Hepatocyte polyploidization and its association with pathophysiological processes. Cell Death Dis. 2017;8(5):e2805.
13. Gentric G, Maillet V, Paradis V, et al. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest. 2015;125(3):981–992.
14. Yamada T, Sogawa K, Kim JK, et al. Increased polyploidy, delayed mitosis and reduced protein phosphatase-1 activity associated with excess copper in the Long Evans Cinnamon rat. Res Commun Mol Pathol Pharmacol. 1998;99(3):283–304.
15. Toyoda H, Bregerie O, Vallet A, et al. Changes to hepatocyte ploidy and binuclearity profiles during human chronic viral hepatitis. Gut. 2005;54(2):297–302.
16. Lin H, Huang YS, Fustin JM, et al. Hyperpolyploidization of hepatocyte initiates preneoplastic lesion formation in the liver. Nat Commun. 2021;12(1):645.
17. Bou-Nader M, Caruso S, Donne R, et al. Polyploidy spectrum: a new marker in HCC classification. Gut. 2020;69(2):355–364.
18. Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J Hepatol. 2015;62(6):1420–1429.
19. Unitt E, Marshall A, Gelson W, et al. Tumour lymphocytic infiltrate and recurrence of hepatocellular carcinoma following liver transplantation. J Hepatol. 2006;45(2):246–253.
20. Gao Q, Qiu SJ, Fan J, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25(18):2586–2593.
21. Berraondo P, Sanmamed MF, Ochoa MC, et al. Cytokines in clinical cancer immunotherapy. Br J Cancer. 2019;120(1):6–15.
22. Pellegrini M, Mak TW, Ohashi PS. Fighting cancers from within: augmenting tumor immunity with cytokine therapy. Trends Pharmacol Sci. 2010;31(8):356–363.
23. Mano Y, Yoshizumi T, Yugawa K, et al. Lymphocyte-to-monocyte ratio is a predictor of survival after liver transplantation for hepatocellular carcinoma. Liver Transpl. 2018;24(11):1603–1611.
24. Xu X, Ling Q, Wang J, et al. Donor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese population. Int J Cancer. 2016;138(3):620–629.
25. Ismael MN, Forde J, Milla E, Khan W, Cabrera R. Utility of inflammatory markers in predicting hepatocellular carcinoma survival after liver transplantation. Biomed Res Int. 2019;2019:7284040.
26. Mazzaferro V, Sposito C, Zhou J, et al. Metroticket 2.0 model for analysis of competing risks of death after liver transplantation for hepatocellular carcinoma. Gastroenterology. 2018;154(1):128–139.
27. Duvoux C, Roudot-Thoraval F, Decaens T, et al. Liver transplantation for hepatocellular carcinoma: a model including alpha-fetoprotein improves the performance of Milan criteria. Gastroenterology. 2012;143(4):
28. Zhang AB, Zhang ZH, Zhang J, et al. Lower mean platelet volume is a risk indicator of hepatocellular carcinoma recurrence following liver transplantation. Hepatobiliary Pancreat Dis Int. 2019;18(3):223–227.
29. Yang Z, Luo FZ, Wang S, et al. Alpha-fetoprotein and (18) F-FDGstandard uptake value predict tumor recurrence after liver transplantation for hepatocellular carcinoma with portal vein tumor thrombosis: preliminary experience. Hepatobiliary Pancreat Dis Int. 2020;19(3):229–234.
30. Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22(13):1166–1175.
31. Tanami S, Ben-Moshe S, Elkayam A, Mayo A, Bahar Halpern K, Itzkovitz S. Dynamic zonation of liver polyploidy. Cell Tissue Res. 2017;368(2):405–410.
32. Xie H, Zhang L, Guo D, et al. Protein profiles of pretransplant grafts predict early allograft dysfunction after liver transplantation from donation after circulatory death. Transplantation. 2020;104(1):79–89.
33. Wu Q, Zhou W, Yin S, et al. Blocking Triggering receptor expressed on myeloid cells-1-positive tumor-associated macrophages induced by hypoxia reverses immunosuppression and anti-programmed cell death ligand 1 resistance in liver cancer. Hepatology. 2019;70(1):198–214.
34. Calderaro J, Rousseau B, Amaddeo G, et al. Programmed death ligand 1 expression in hepatocellular carcinoma: relationship with clinical and pathological features. Hepatology. 2016;64(6):2038–2046.
35. Huang CY, Wang Y, Luo GY, et al. Relationship between PD-L1 expression and CD8+ T-cell immune responses in Hepatocellular carcinoma. J Immunother. 2017;40(9):323–333.
36. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228.
37. Bielski CM, Zehir A, Penson AV, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet. 2018;50(8):1189–1195.
38. Wangsa D, Quintanilla I, Torabi K, et al. Near-tetraploid cancer cells show chromosome instability triggered by replication stress and exhibit enhanced invasiveness. FASEB J. 2018;32(7):3502–3517.
39. Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140.
40. Lee IC, Huang YH, Chau GY, et al. Serum interferon gamma level predicts recurrence in hepatocellular carcinoma patients after curative treatments. Int J Cancer. 2013;133(12):2895–2902.
41. Senovilla L, Vitale I, Martins I, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337(6102):1678–1684.
42. Aranda F, Chaba K, Bloy N, et al. Immune effectors responsible for the elimination of hyperploid cancer cells. Oncoimmunology. 2018;7(8):e1463947.
43. Lopez-Soto A, Gonzalez S, Lopez-Larrea C, Kroemer G. Immunosurveillance of malignant cells with complex karyotypes. Trends Cell Biol. 2017;27(12):880–884.
44. Lin YH, Zhang S, Zhu M, et al. Mice with increased numbers of polyploid hepatocytes maintain regenerative capacity but develop fewer hepatocellular carcinomas following chronic liver injury. Gastroenterology. 2020;158(6):1698–1712e1614.
45. Sladky VC, Knapp K, Szabo TG, et al. PIDDosome-induced p53-dependent ploidy restriction facilitates hepatocarcinogenesis. EMBO Rep. 2020;21(12):e50893.
46. Zhang S, Zhou K, Luo X, et al. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell. 2018;44(4):447–459e445.
47. Zhang S, Nguyen LH, Zhou K, et al. Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology. 2018;154(5):1421–1434.
48. Gjelsvik KJ, Besen-McNally R, Losick VP. Solving the polyploid mystery in health and disease. Trends Genet. 2019;35(1):6–14.
49. Kucukoglu O, Sowa JP, Mazzolini GD, Syn WK, Canbay A. Hepatokines and adipokines in NASH-related hepatocellular carcinoma. J Hepatol. 2021;74(2):442–457.
50. Firl DJ, Sasaki K, Agopian VG, et al. Charting the path forward for risk prediction in liver transplant for hepatocellular carcinoma: international validation of HALTHCC among 4089 patients. Hepatology. 2020;71(2):569–582.
51. Cucchetti A, Serenari M, Sposito C, et al. Including mRECIST in the Metroticket 2.0 criteria improves prediction of hepatocellular carcinoma-related death after liver transplant. J Hepatol. 2020;73(2):342–348.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.