Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 12
Polymorphism Of The APM1 Gene In Subjects With Central Obesity Related To Lower High-Density Lipoprotein Cholesterol
Authors Barliana MI , Yolanda PD, Rostinawati T , Ng H , Alfian SD , Abdulah R , Diantini A
Received 19 June 2019
Accepted for publication 16 October 2019
Published 6 November 2019 Volume 2019:12 Pages 2317—2324
DOI https://doi.org/10.2147/DMSO.S220050
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Juei-Tang Cheng
Melisa I Barliana,1,2 Poppy D Yolanda,1 Tina Rostinawati,1 Henry Ng,1 Sofa D Alfian,2,3 Rizky Abdulah,2,3 Ajeng Diantini2,3
1Department of Biological Pharmacy, Biotechnology Laboratory, Faculty of Pharmacy, Universitas Padjadjaran, Bandung, Indonesia; 2Center of Excellence in Higher Education for Pharmaceutical Care Innovation, Universitas Padjadjaran, Bandung, Indonesia; 3Department of Pharmacology and Clinical Pharmacy, Universitas Padjadjaran, Bandung, Indonesia
Correspondence: Melisa I Barliana
Department of Biological Pharmacy, Biotechnology Laboratory, Faculty of Pharmacy, Universitas Padjadjaran, Jl. Raya Bandung Sumedang KM. 21, Bandung, Jatinangor 45363, Indonesia
Tel +62 22 8428 8888 Ext 3510 Email [email protected]
Background: Central obesity is a risk factor for metabolic syndrome. Subjects with central obesity have a higher risk of developing type 2 diabetes and cardiovascular disease. Many conditions affect the prevalence of central obesity, including energy expenditure, aging, proinflammatory conditions, and hormonal, genetic, and ethnic differences. Polymorphism of the APM1 gene, encoding the protein adiponectin, is closely related to metabolic syndrome. Adiponectin influences fatty acid oxidation and glucose intake in muscle. Therefore, variation in the APM1 gene is associated with diabetes and obesity.
Purpose: The aim of the present study was to investigate the correlation of the single-nucleotide polymorphism (SNP) of the APM1 SNP rs2241766 with body mass index (BMI) and lipid profiles in Indonesian (Bandung) subjects.
Patients and methods: Genotyping of the APM1 gene was performed using the Amplification Refractory Mutation System. Whole blood and serum of 54 subjects with central obesity (waist circumference [WC] ≥90 cm) and 53 healthy subjects (WC Results: The TT and GT genotype were observed (no GG genotype) in all subjects. The TC, LDL, fasting blood glucose, and BMI did not show a significant correlation between genotype variations of APM1 with central obesity. Otherwise, subjects with central obesity with the TT genotype had lower HDL levels than those with the GT genotype (p = 0.014, significant OR 1.045; 95% CI).
Conclusion: This finding suggests that the T allele of the APM1 SNP rs2241766 is dominant in the Bandung population, and subjects with the homozygous TT genotype have a higher incidence of metabolic disorder.
Keywords: waist circumference, SNP, rs2241766, adiponectin, metabolic syndrome
Introduction
Central obesity, or abdominal visceral obesity, has become a major risk factor for insulin resistance, type 2 diabetes mellitus (T2DM), hypertension, and dyslipidemia, which are linked to metabolic disorders and cardiovascular disease.1–8 Central obesity is a consequence of excessive visceral adipose tissue (VAT) accumulation, which is correlated with a high mortality rate. It was also observed as ectopic triglyceride storage and a source of proinflammatory cytokines.4,9 Measurement of central obesity by waist circumference (WC) was shown to be a better predictor of MetS than Body Mass Index (BMI), the waist-to-hip ratio, and waist-to-height ratio.10
Adipose tissue secretes many cytokines, called adipokines, such as leptin, adiponectin, tumor necrosis factor alpha (TNFα), Interleukin-1β (IL-1β), and Interleukin-6 (IL-6), which are linked to chronic low-grade inflammation and the inflammatory response. These proinflammatory adipokines play an important role in energy metabolism. They may modify carbohydrate and lipid metabolism, resulting in a higher risk of insulin resistance, MetS, and cardiovascular diseases.9,11–13
Adiponectin is a hormone encoded by the Adipose Most Abundant Gene Transcript 1 (APM1) gene and secreted by fat cells in adipose tissue.14 Adiponectin consists of 3 exons spanning 16 kb, and APM1 is localized on the long arm of chromosome 3 in the 3q27 region.13,15 Adiponectin regulates carbohydrate and lipid metabolism by decreasing hepatic gluconeogenesis, stimulating glucose uptake, and enhancing insulin sensitivity.16 Adiponectin may also inhibit monocyte and macrophage adhesion to endothelial cells.13,17–19 Therefore, adiponectin has anti-inflammatory, antidiabetic, anti-atherogenic, and cardioprotective properties.13,16,20,21 Low levels of adiponectin are observed in obese,13,21,22 centrally obese and Mets subjects that will affect their overall health.7,23,24 Increased VAT will increase the size of fat cell but is negatively correlated with adiponectin serum level.25 Decreased serum adiponectin levels were positively correlated with increased total cholesterol, low-density lipoprotein (LDL) cholesterol, and small, dense LDL particles in T2DM subjects.26
Abnormal adiponectin secretion is also affected by Single-Nucleotide Polymorphisms (SNPs) of the APM1 gene but inconsistency was observed across populations.27 The most commonly studied SNPs in the APM1 gene are rs2241766 (T45G) and rs1501299 (G276T).3,28,29 The APM1 gene polymorphism is associated with obesity,30 and both rs2241766 (T45G) and rs1501299 (G276T) were found to affect adiponectin secretion.29,31 The rs2241776 polymorphism is a silent T to G substitution mutation in exon 2 that is correlated with an increased risk of diabetic nephropathy in T2DM and decreased serum levels of adiponectin.29,32
In Indonesia, the prevalence of MetS increases every year, in step with increased central obesity and/or BMI.33 The risk of obesity is affected by several factors, including the environment and genetics.34 Many studies conducted in other countries showed correlations between obesity, BMI, WC, or lipid profiles with pharmacogenomic or genetic variations. However, there are very limited data for genomic studies, especially those on disease-related SNPs, on the Indonesian population. Therefore, the aim of present study was to investigate the correlation of APM1 genotype variation (rs2241776) with BMI and lipid profiles (total, high-density lipoprotein [HDL], and LDL cholesterol) in subjects with central obesity in Indonesia, especially the Bandung city population. The results will provide a first report describing variation in the APM1 gene (rs2241776) in subjects with central obesity, which can be used to compare the Bandung population with other populations and to pinpoint the potential role of genetics and fat distribution in ethnic differences in health outcomes.
Materials And Methods
Subjects
A case-control study of 107 subjects (54 with central obesity and 53 controls) in Bandung city was conducted to investigate the correlation of the SNP rs2241766 in the APM1 gene with lipid profiles and BMI. All subjects were men, >18 years old, with WC <90 cm (for control subjects) and ≥90 cm (for subjects with central obesity).8,33,35,36 The patient population who meet the criteria for inclusion was selected randomly from the general population that attended one laboratory clinic in Bandung city for routine examinations.
All participant information was anonymized during the analyses, and all patients gave written informed consent to participate. All procedures performed in studies involving human participants complied with the ethical standards of the institutional and/or national research committee, Medical Research Ethics Committee of the Medical Faculty at Universitas Padjadjaran (No. 824/UN6.C1.3.2/KEPK/PN/2015). The present research was also conducted in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Anthropometric Measurements
WC, weight, and height were measured for all subjects. WC was measured at the narrowest point between the lowest rib and the iliac crest. BMI was calculated as the weight (kg) divided by the height (m2).35
Biochemical Parameters For Metabolic Disorders
Serum samples from overnight fasted (≥10 h) subjects were analyzed for lipid profiles (total, HDL, and LDL cholesterol) and blood glucose. The criteria for subjects with metabolic disorders were BMI >25 kg/m2, total cholesterol (TC) > 200 mg/dL, LDL > 100 mg/dL, HDL < 40 mg/dL, and fasting blood glucose (FBG) >110 mg/dL.8,33 All parameters were measured by Abbot Architect c4000 (Abbot, Abbot Park, USA) in a clinical laboratory with closed system reagents. HDL and LDL were measured using homogenous enzymatic assay, total cholesterol using CHOD-PAP, FBG using hexokinase method, and triglyceride using GPO-PAP.
Genotyping Of APM1 rs2241766
Information on the APM1 variant rs2241766 was obtained from The National Centre for Biotechnology Information (https://www.ncbi.nlm.nih.gov). Samples of whole blood were collected from all groups and stored at −80°C until the assay. Genomic DNA was isolated using the Gene JET Genomic DNA Purification Kit (Thermo Fischer Scientific, Waltham, MA, USA), and PCR was conducted using PCR Master Mix (Thermo Fischer Scientific) with a specific tetra primer to identify SNPs in the APM1 SNP rs2241766. Tetra primers were designed using the software GENETYX version 10 and confirmed by Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html). The tetra primers used in this study were as follows: forward primer 1 (F1) 5′-TGTGCTTGGTCCTGTGCTCAGACA-3′; forward primer 2 (F2) 5′-TTCTACTGCTATTAGCTCTGCCCGGG-3′; reverse primer 1 (R1) 5′-CTTGA GTCGTGGTTTCCTGGTCATGA-3′; and reverse primer 2 (R2) 5′- ACCAACAGAGCCTTGCACATTAGAC-3′ (Sigma-Aldrich Co., St Louis, MO, USA). PCR fragments were electrophoresed through a 2% agarose gel and visualized under ultraviolet light at 312 nm. The following three bands were obtained: control (950 bp), G (613 bp), and T (398 bp) alleles. The internal control for PCR was GAPDH gene expression in the same samples.
Statistical Analysis
Analysis of allele frequencies at each locus was reported using descriptive statistics with Hardy–Weinberg equilibrium (df = 1). Mann–Whitney tests were used to assess the associations of each profile lipid, fasting blood glucose, or body measurement variables with the APM1 genetic variants. Odds ratios (ORs) with a 95% confidence interval were obtained using binary logistic regression analysis. Statistically significant differences of BMIs were determined by Student’s t-test. A p value less than 0.05 was considered statistically significant.
Results
Genotyping Of APM1 rs2241766 In Controls And Subjects With Central Obesity
The PCR fragments using tetra primers were the wild type TT and heterozygote GT genotypes that observed in both groups but not the GG genotype (double mutant) (Figure 1). In the central obesity group (n=54), 32 subjects (59.26%) with the TT genotype and 22 subjects (40.74%) with the GT genotype were observed, whereas in the control group (n=53), 27 (50.94%) and 26 (49.6%) subjects with the TT and GT genotypes, respectively, were observed (Table 1). Statistical analysis showed that there was no correlation between WC and genotype variation (p = 0.387). The frequencies of APM1 SNP rs2241766 genotypes were in a disequilibrium state with p < 0.05 (p = 0.003) according to the Hardy–Weinberg equation (Table 2).
 |
Table 1 APM1 SNP rs2241766 Variations In Central Obesity And Control Subjects (p < 0.05) |
 |
Table 2 Hardy–Weinberg Equilibrium For The Observed And Expected Genotype Frequencies For APM1 SNP rs2241766 |
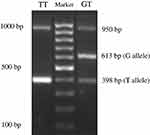 |
Figure 1 Visualization of the APM1 SNP rs2241766 using agarose gel electrophoresis. |
Correlations Between Genotype Variants Of APM1 rs2241766, BMI, And Biochemical Parameters In Both Groups
Majority of the subjects with central obesity had high TC and LDL but low HDL (Table 3). However, some of the control subjects also showed a similar lipid profile. In both groups, only 6 obese and 3 control subjects were in a hyperglycemic state (Table 4). The data showed that obese subjects (28.36±1.34) had higher average of BMIs (>25 kg/m2) than control subjects (19.35±0.06) significantly (p=0.00), in concordance with a larger WC (>90 cm). In obese central group, a lower HDL value showed a significant correlation (p = 0.014, significant OR 1.045; 95% CI) with the TT genotype (Table 4). Total cholesterol (p = 0.306), LDL (p = 0.880), FBG (p = 0.240), and BMI (p = 0.138) have no significant correlation with genotype variations of APM1 in central obesity subjects.
 |
Table 3 Lipid Profiles, FBG And BMI Measurement In Central Obesity And Control Subjects |
 |
Table 4 Lipid Profiles, FBG And BMI Measurement And The Genotype Distribution Of The APM1 SNP rs2241766 In Control And Obese Subjects |
Discussion
The present research is the first study to investigate the correlation of genotype variation in the APM1 gene with BMI and lipid profiles in subjects with central obesity in Indonesia, especially the Bandung city population. Our results showed that the T allele (GT and TT genotype) of APM1 rs2241766, but not the GG genotype, was distributed among controls and the majority of subjects with central obesity. The G allele of APM1 (GT) in control subjects (without central obesity) was not showing significant difference with central obesity subjects. This finding was in line with a study in Belgian Caucasian women, which found that the G allele of rs2241766 was observed at a higher frequency in lean than obese women and was therefore proposed to confer a protective effect against obesity.30 However, another study in a different population reported that the APM1 SNP rs2241766 (GGA haplotype) contributed to the genetic risk of T2DM in a diabetic Japanese Brazilian population.28 The T to G variant was significantly correlated with the risk of T2DM in non-diabetic Japanese, Chinese Han, and Turkish populations,31,37,38 insulin resistance in a Korean population,39,40 and the risk of hyperglycemia41 and diabetic peripheral neuropathy in a Chinese population.29
In the present study, subjects with central obesity had higher BMIs than lean subjects (49 of 54 subjects with central obesity). The majority of subjects with central obesity had high BMIs, although we also observed a small number of lean subjects with high BMIs (4 subjects). Subjects with central obesity and a high BMI had a high risk of MetS and cardiovascular disease. These findings are concordant with those of a previous study that showed BMI was correlated with major risk factors for cardiometabolic disease and death in the USA but less sensitive and specific for predicting obesity-related health problems.10 Moreover, Asian populations have a low prevalence of obesity according to BMI parameters than other ethnic groups. WC was used to measure central obesity, which is a sensitive indicator of fat distribution correlated with MetS.10,42–45 In Asians, a lower BMI is associated with a higher amount of visceral fat than in Hispanic or white people. This explains why dyslipidemia, diabetes, and hypertension have a high prevalence in Asian populations.43,46
Central obesity is characterized by a low-grade chronic inflammation that contributes to hypertension, hypercholesterolemia, low HDL, and hyperglycaemia.8,31 Therefore, central obesity is strongly correlated with mortality caused by cardiovascular disease.47 Another study showed that normal weight subjects with central obesity have a higher mortality risk, specifically 87% higher, compared to subjects with similar BMIs without central obesity.48 Similarly, we observed that subjects with central obesity had a high level of serum TC and LDL and a low level of HDL. Hyperglycaemia was not observed in our study, even in subjects with central obesity.
HDL has been associated with cardiometabolic health,49–51 although recent studies reported that the cardioprotective effect of HDL also depends on HDL subclasses,52 body fat distribution,53 and BMI.54 In our study, most of the subjects with central obesity had the TT genotype of the APM1 SNP rs2241766 and also showed higher levels of TC and LDL but lower HDL. Furthermore, the lipid profiles of subjects with central obesity showed higher levels of TC and LDL (although not significant) and low HDL levels. These findings show that people with the T allele are more susceptible to central obesity. Subjects with central obesity with the TT genotype have no G allele, which has a protective effect against becoming obese.30 But in the Bandung city population, subjects with the GT genotype were observed to have low HDL levels. So, the G allele was not shown to have a protective effect against cardiovascular disease, considering the low level of HDL in the population with the GT genotype. This discrepancy might be one of the limitations of the present study, which was conducted on a small segment of a single population.
In a Chinese population, it was shown that a reduced serum adiponectin concentration was correlated with reduced HDL cholesterol and increased MetS risk, which is positively correlated with central obesity. In a Chinese population, the frequency of the GG genotype of the APM1 SNP rs2241766 was higher in the MetS group than that of the TT and GT genotypes; moreover, the GG genotype was a significant independent risk factor for MetS.55 The level of adiponectin in serum showed a dependence on genotype variation of the APM1 SNP rs2241766. The G allele was associated with lower serum adiponectin levels in subjects with high BMIs.56 This report contradicts our results, although we did not find the GG genotype in our samples. We observed that the TT genotype was found in most of the subjects with central obesity, whereas the GT genotype was found most in lean subjects. In the present study, we did not measure adiponectin levels in serum. So, we cannot confirm whether variation of the APM1 SNP rs2241766 in the Bandung city population affects the concentration and expression of adiponectin.
Variation in the APM1 gene has been associated variably with increases in BMI, insulin resistance, and T2DM.16,57 These variations are also correlated with cancer because of its function in glucose and lipid metabolism. In a Mexican population with ductal infiltrating breast cancer (DIBC), patients with the TT and GT genotypes of the APM1 SNP (rs2241766) showed higher values of WC, hip circumference (HP), and waist-hip ratio (WHR).58
Genetic variation might be applied in personalized medicine, as it is related to pharmacogenomics but highly specific in a population. Different geographical regions or even subpopulations, might show different allele frequencies of SNPs.59 SNPs related to personalized medicine affect the practice of medicine, as they are predictive biomarkers for patient stratification and dose selection or adjustment.60 Pharmacogenomic research in Asian regions is still limited compared with Western countries, of which the pioneering countries are Japan, Korea, Singapore, Thailand, and India.59 This condition is also observed in variations of the APM1 SNP rs2241766. Therefore, data from pharmacogenomic studies, especially those on disease-related SNPs, on the Indonesian population are very limited.
Conclusion
By understanding the correlation between the APM1 gene SNP rs2241766, WC, and lipid profiles, will be a potential parameter for MetS risk in Indonesian population. In our study, the T allele was observed at a high frequency in subjects with central obesity with high levels of LDL and TC. The GT genotype was associated with a lower level of HDL in subjects with central obesity. These data were discordant with other reports. Even though the G allele had a protective effect against obesity, it was not protective against cardiovascular disease. However, adiponectin measurement in serum is needed to complete the analysis to confirm that genetic variation of the APM1 SNP rs2241766 in Indonesia affects the expression and function of adiponectin. Furthermore, more samples are needed for further study in the Indonesian population, considering that Indonesia has many tribes spread out on different islands. Despite its limitations, the present study is the first study of variation in the APM1 SNP rs2241766 in Indonesia in correlation with central obesity, BMI, lipid profiles, and FBG.
Acknowledgment
This research was funded by Academic Leadership Grant Universitas Padjadjaran for AD.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–846. doi:10.1038/nature05482
2. Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obesity Rev. 2010;11(1):11–18. doi:10.1111/obr.2010.11.issue-1
3. Cao D, Ouyang S, Liu Z, Ma F, Wu J. Association of the ADIPOQ T45G polymorphism with insulin resistance and blood glucose: a meta-analysis. Endocr J. 2014;61(5):437–446. doi:10.1507/endocrj.EJ13-0444
4. Grundy SM. Adipose tissue and metabolic syndrome: too much, too little or neither. Eur J Clin Invest. 2015;45(11):1209–1217. doi:10.1111/eci.12519
5. Wijaya CR, Sukmana IR. J L. predictor of diabetes: correlation between leucine concentration and insulin resistance. Pharmacol Clin Pharm Res. 2017;2(3):70–74. doi:10.15416/pcpr.v2i3.16225
6. Grundy SM, Cleeman JI, Daniels SR, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute scientific statement: executive Summary. Crit Pathw Cardiol. 2005;4(4):198–203. doi:10.1097/00132577-200512000-00018
7. Ntzouvani A, Fragopoulou E, Panagiotakos D, Pitsavos C, Antonopoulou S. Reduced circulating adiponectin levels are associated with the metabolic syndrome independently of obesity, lipid indices and serum insulin levels: a cross-sectional study. Lipids Health Dis. 2016;15(1):140. doi:10.1186/s12944-016-0311-7
8. Federation ID. The IDF consensus worldwide definition of the metabolic syndrome. IDF Commun. 2006;(February):1–23.
9. Tchernof A, Despres JP. Pathophysiology of human visceral obesity: an update. Physiol Rev. 2013;93(1):359–404. doi:10.1152/physrev.00033.2011
10. Bener A, Yousafzai MT, Darwish S, Al-Hamaq AO, Nasralla EA, Abdul-Ghani M. Obesity index that better predict metabolic syndrome: body mass index, waist circumference, waist hip ratio, or waist height ratio. J Obes. 2013;2013:269038. doi:10.1155/2013/269038
11. Trayhurn P, Wood IS. Signalling role of adipose tissue: adipokines and inflammation in obesity. Biochem Soc Trans. 2005;33(Pt 5):1078–1081. doi:10.1042/BST0331078
12. Gateva A, Assyov Y, Tsakova A, Kamenov Z. Classical (adiponectin, leptin, resistin) and new (chemerin, vaspin, omentin) adipocytokines in patients with prediabetes. Horm Mol Biol Clin Investig. 2018;34:1.
13. Nigro E, Scudiero O, Monaco ML, et al. New insight into adiponectin role in obesity and obesity-related diseases. Biomed Res Int. 2014;2014:658913. doi:10.1155/2014/658913
14. Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose most abundant gene transcript 1). Biochem Biophys Res Commun. 1996;221(2):286–289. doi:10.1006/bbrc.1996.0587
15. Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11(21):2607–2614. doi:10.1093/hmg/11.21.2607
16. Rabe K, Lehrke M, Parhofer KG, Broedl UC. Adipokines and insulin resistance. Mol Med. 2008;14(11–12):741–751. doi:10.2119/2008-00058.Rabe
17. Ouchi N, Kihara S, Arita Y, et al. Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation. 1999;100(25):2473–2476. doi:10.1161/01.CIR.100.25.2473
18. Subedi A, Park PH. Autocrine and paracrine modulation of microRNA-155 expression by globular adiponectin in RAW 264.7 macrophages: involvement of MAPK/NF-kappaB pathway. Cytokine. 2013;64(3):638–641. doi:10.1016/j.cyto.2013.09.011
19. Nigro E, Scudiero O, Sarnataro D, et al. Adiponectin affects lung epithelial A549 cell viability counteracting TNFalpha and IL-1ss toxicity through AdipoR1. Int J Biochem Cell Biol. 2013;45(6):1145–1153. doi:10.1016/j.biocel.2013.03.003
20. Lee B, Shao J. Adiponectin and energy homeostasis. Rev Endocr Metab Disord. 2014;15(2):149–156. doi:10.1007/s11154-013-9283-3
21. Ohashi K, Ouchi N, Matsuzawa Y. Anti-inflammatory and anti-atherogenic properties of adiponectin. Biochimie. 2012;94(10):2137–2142. doi:10.1016/j.biochi.2012.06.008
22. Bahia L, Aguiar LG, Villela N, et al. Relationship between adipokines, inflammation, and vascular reactivity in lean controls and obese subjects with metabolic syndrome. Clinics (Sao Paulo). 2006;61(5):433–440. doi:10.1590/S1807-59322006000500010
23. Halleux CM, Takahashi M, Delporte ML, et al. Secretion of adiponectin and regulation of apM1 gene expression in human visceral adipose tissue. Biochem Biophys Res Commun. 2001;288(5):1102–1107. doi:10.1006/bbrc.2001.5904
24. Moon HU, Ha KH, Han SJ, Kim HJ, Kim DJ. The association of adiponectin and visceral fat with insulin resistance and beta-cell dysfunction. J Korean Med Sci. 2019;34(1):e7. doi:10.3346/jkms.2019.34.e7
25. Meyer LK, Ciaraldi TP, Henry RR, Wittgrove AC, Phillips SA. Adipose tissue depot and cell size dependency of adiponectin synthesis and secretion in human obesity. Adipocyte. 2013;2(4):217–226. doi:10.4161/adip.24953
26. Eslamian M, Mohammadinejad P, Aryan Z, Nakhjavani M, Esteghamati A. Positive correlation of serum adiponectin with lipid profile in patients with type 2 diabetes mellitus is affected by metabolic syndrome status. Arch Iran Med. 2016;19(4):269–274.
27. Wu J, Liu Z, Meng K, Zhang L. Association of adiponectin gene (ADIPOQ) rs2241766 polymorphism with obesity in adults: a meta-analysis. PLoS One. 2014;9(4):e95270. doi:10.1371/journal.pone.0095270
28. Vendramini MF, Pereira AC, Ferreira SR, Kasamatsu TS, Moises RS. Association of genetic variants in the adiponectin encoding gene (ADIPOQ) with type 2 diabetes in Japanese Brazilians. J Diabetes Complications. 2010;24(2):115–120. doi:10.1016/j.jdiacomp.2009.01.007
29. Ji ZY, Li HF, Lei Y, et al. Association of adiponectin gene polymorphisms with an elevated risk of diabetic peripheral neuropathy in type 2 diabetes patients. J Diabetes Complications. 2015;29(7):887–892. doi:10.1016/j.jdiacomp.2015.06.008
30. Beckers S, Peeters AV, de Freitas F, et al. Association study and mutation analysis of adiponectin shows association of variants in APM1 with complex obesity in women. Ann Hum Genet. 2009;73(Pt 5):492–501. doi:10.1111/j.1469-1809.2009.00532.x
31. Tu Y, Yu Q, Fan G, et al. Assessment of type 2 diabetes risk conferred by SNPs rs2241766 and rs1501299 in the ADIPOQ gene, a case/control study combined with meta-analyses. Mol Cell Endocrinol. 2014;396(1–2):1–9. doi:10.1016/j.mce.2014.08.006
32. Gibson F, Froguel P. Genetics of the APM1 locus and its contribution to type 2 diabetes susceptibility in French Caucasians. Diabetes. 2004;53(11):2977–2983. doi:10.2337/diabetes.53.11.2977
33. Indonesia MoHRo. Riset Kesehatan Dasar 2013. Badan Penelitian dan Pengembangan Kesehatan, Ministry of Health Republic Indonesia 2013:1–384.
34. Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20.
35. Consultation W. Waist circumference and waist-hip ratio. Report WHO Expert Consultation Geneva. 2008;2008:8–11.
36. Nishida C, Ko GT, Kumanyika S. Body fat distribution and noncommunicable diseases in populations: overview of the 2008 WHO expert consultation on waist circumference and Waist-Hip Ratio. Eur J Clin Nutr. 2010;64(1):2–5. doi:10.1038/ejcn.2009.139
37. Nakatani K, Noma K, Nishioka J, et al. Adiponectin gene variation associates with the increasing risk of type 2 diabetes in non-diabetic Japanese subjects. Int J Mol Med. 2005;15(1):173–177.
38. Arikoglu H, Ozdemir H, Kaya DE, et al. The adiponectin variants contribute to the genetic background of type 2 diabetes in Turkish population. Gene. 2014;534(1):10–16. doi:10.1016/j.gene.2013.10.039
39. Jang Y, Lee JH, Chae JS, et al. Association of the 276G->T polymorphism of the adiponectin gene with cardiovascular disease risk factors in nondiabetic Koreans. Am J Clin Nutr. 2005;82(4):760–767. doi:10.1093/ajcn/82.4.760
40. Shin MJ, Jang Y, Koh SJ, et al. The association of SNP276G>T at adiponectin gene with circulating adiponectin and insulin resistance in response to mild weight loss. Int J Obes (Lond). 2006;30(12):1702–1708. doi:10.1038/sj.ijo.0803338
41. Tso AW, Sham PC, Wat NM, et al. Polymorphisms of the gene encoding adiponectin and glycaemic outcome of Chinese subjects with impaired glucose tolerance: a 5-year follow-up study. Diabetologia. 2006;49(8):1806–1815. doi:10.1007/s00125-006-0324-2
42. Rankinen T, Kim SY, Perusse L, Despres JP, Bouchard C. The prediction of abdominal visceral fat level from body composition and anthropometry: ROC analysis. Int J Obesity Related Metab Disord. 1999;23(8):801–809. doi:10.1038/sj.ijo.0800929
43. Liu X, Chen Y, Boucher NL, Rothberg AE. Prevalence and change of central obesity among US Asian adults: NHANES 2011-2014. BMC Public Health. 2017;17(1):678. doi:10.1186/s12889-017-4689-6
44. Zaninotto P, Pierce M, Breeze E, de Oliveira C, Kumari M. BMI and waist circumference as predictors of well-being in older adults: findings from the English longitudinal study of ageing. Obesity (Silver Spring). 2010;18(10):1981–1987. doi:10.1038/oby.2009.497
45. Hendra P, Virginia DM, Fenty F, Widayati A. Correlation between anthropometric measurement and lipid profile among rural community at Cangkringan Village, District Sleman, Yogyakarta Province. Indonesian J Clin Pharm. 2017;6(2):107–115. doi:10.15416/ijcp
46. Misra A, Khurana L. Obesity and the metabolic syndrome in developing countries. J Clin Endocrinol Metab. 2008;93(11 Suppl 1):S9–30. doi:10.1210/jc.2008-1595
47. Coutinho T, Goel K, Correa de Sa D, et al. Combining body mass index with measures of central obesity in the assessment of mortality in subjects with coronary disease: role of “normal weight central obesity”. J Am Coll Cardiol. 2013;61(5):553–560. doi:10.1016/j.jacc.2012.10.035
48. Sahakyan KR, Somers VK, Rodriguez-Escudero JP, et al. Normal-weight central obesity: implications for total and cardiovascular mortality. Ann Intern Med. 2015;163(11):827–835. doi:10.7326/M14-2525
49. Khera AV, Demler OV, Adelman SJ, et al. Cholesterol efflux capacity, high-density lipoprotein particle number, and incident cardiovascular events: an analysis From the JUPITER trial (Justification for the use of statins in prevention: an intervention trial evaluating rosuvastatin). Circulation. 2017;135(25):2494–2504. doi:10.1161/CIRCULATIONAHA.116.025678
50. Verdoia M, Schaffer A, Suryapranata H, De Luca G. Effects of HDL-modifiers on cardiovascular outcomes: a meta-analysis of randomized trials. Nutr Metab Cardiovasc Dis. 2015;25(1):9–23. doi:10.1016/j.numecd.2014.09.003
51. Iskandar MF, Nurmainah N, Susanti R. The risk of atherosclerotic cardiovascular disease among hyperlipidaemia patients and appropriateness of statin therapy. Pharmacol Clin Pharm Res. 2018;3(1):21–25. doi:10.15416/pcpr.
52. Woudberg NJ, Lecour S, Goedecke JH. HDL subclass distribution shifts with increasing central adiposity. J Obes. 2019;2019:2107178. doi:10.1155/2019/2107178
53. Goedecke JH, Levitt NS, Lambert EV, et al. Differential effects of abdominal adipose tissue distribution on insulin sensitivity in black and white South African women. Obesity (Silver Spring). 2009;17(8):1506–1512. doi:10.1038/oby.2009.73
54. Woudberg NJ, Goedecke JH, Blackhurst D, et al. Association between ethnicity and obesity with high-density lipoprotein (HDL) function and subclass distribution. Lipids Health Dis. 2016;15:92. doi:10.1186/s12944-016-0257-9
55. Li P, Jiang R, Li L, Liu C, Yang F, Qiu Y. Correlation of serum adiponectin and adiponectin gene polymorphism with metabolic syndrome in Chinese adolescents. Eur J Clin Nutr. 2015;69(1):62–67. doi:10.1038/ejcn.2014.152
56. Pollin TI, Tanner K, O’Connell JR, et al. Linkage of plasma adiponectin levels to 3q27 explained by association with variation in the APM1 gene. Diabetes. 2005;54(1):268–274. doi:10.2337/diabetes.54.1.268
57. Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56(5):1198–1209. doi:10.2337/db06-0506
58. Macias-Gomez NM, Hernandez-Terrones MC, Ramirez-Guerrero AA, Leal-Ugarte E, Gutierrez-Angulo M, Peregrina-Sandoval J. ADIPOQ rs2241766 SNP as protective marker against DIBC development in Mexican population. PLoS One. 2019;14(3):e0214080. doi:10.1371/journal.pone.0214080
59. Ang HX, Chan SL, Sani LL, et al. Pharmacogenomics in Asia: a systematic review on current trends and novel discoveries. Pharmacogenomics. 2017;18(9):891–910. doi:10.2217/pgs-2017-0009
60. Laing RE, Hess P, Shen Y, Wang J, Hu SX. The role and impact of SNPs in pharmacogenomics and personalized medicine. Curr Drug Metab. 2011;12(5):460–486. doi:10.2174/138920011795495268
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
