Back to Journals » OncoTargets and Therapy » Volume 13
Plasma Exosomal Long Noncoding RNA lnc-SLC2A12-10:1 as a Novel Diagnostic Biomarker for Gastric Cancer
Authors Zheng P , Zhang H, Gao H, Sun J, Li J, Zhang X , Gao L, Ma P, Li S
Received 12 March 2020
Accepted for publication 23 April 2020
Published 11 May 2020 Volume 2020:13 Pages 4009—4018
DOI https://doi.org/10.2147/OTT.S253600
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Peiming Zheng,1,* Haoliang Zhang,2,* Huijie Gao,3,* Jingfang Sun,2 Junmeng Li,4 Xiulei Zhang,5 Lan Gao,1 Ping Ma,2,6 Shibao Li2,6
1Department of Clinical Laboratory, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, People’s Republic of China; 2Department of Laboratory Medicine, Affiliated Hospital of Xuzhou Medical University, Xuzhou 221002, People’s Republic of China; 3Department of Oncology, The First Affiliated Hospital of Henan University, Kaifeng 450001, People’s Republic of China; 4Department of Gastrointestinal Surgery, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, People’s Republic of China; 5Department of Microbiome Laboratory, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou 450003, People’s Republic of China; 6Medical Technology School of Xuzhou Medical University, Xuzhou 221004, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lan Gao; Ping Ma Email [email protected]; [email protected]
Purpose: Exosomes participate in cellular communications by transmitting active molecules, including long noncoding RNAs (lncRNAs) and are regarded as suitable candidates for disease diagnosis. This study aimed to identify gastric cancer (GC)-specific exosomal lncRNA and investigate the potential diagnostic value of plasma exosomal lncRNA in GC.
Patients and Methods: Exosomes from the culture media (CM) of four GC cells (GCCs) and human gastric epithelial cells were isolated. Exosomal RNA was extracted, and lncRNA microarray assay was performed to identify GC-specific exosomal lncRNAs. The expression levels of the candidate exosomal lncRNAs were validated in 120 subjects via quantitative reverse transcription PCR (qRT-PCR). The receiver operating characteristic (ROC) curve and area under curve were used to estimate the diagnostic capacity. We investigated the potential relationship between plasma exosomal lncRNA expression and the clinicopathological parameters of GC.
Results: A total of 199 exosomal lncRNAs were expressed at considerable higher levels in GCCs than those in normal controls, among which the top 10 upregulated lncRNAs were selected for further validation in cell, CM, and plasma. qRT-PCR revealed that lnc-SLC2A12-10:1 was remarkably upregulated in exosomes derived from patients with GC and GCCs. The area under the ROC curve was 0.776, which was higher than the diagnostic accuracies of CEA, CA 19– 9, and CA72– 4. The expression level of exosomal lnc-SLC2A12-10:1 was also significantly correlated with tumor size, TNM stage, lymph node metastasis, and degree of differentiation. The postoperative expression levels of exosomal lnc-SLC2A12-10:1 were lower compared with those of preoperative levels.
Conclusion: Our study suggested that exosomal lnc-SLC2A12-10:1 may be a potential noninvasive biomarker for the diagnosis and prognosis monitoring of GC. Further large-scale studies are necessary to validate its performance in GC progression.
Keywords: exosomes, biomarker, gastric cancer, long noncoding RNA, diagnosis
Introduction
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer-related death worldwide, with its highest incidence rate recorded in East Asia.1,2 In China, GC remains the second most prevalent cancer, with approximately 679,100 new cases and 498,000 deaths in 2015.3 Most patients are diagnosed at the advanced stage, and their 5-year overall survival rates are less than 25% due to the lack of sensitive and specific early diagnosis.4 Therefore, novel and reliable GC-specific biomarkers should be developed for early GC diagnosis.
Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs with a length longer than 200 nucleotides and limited protein-coding potential.5 Emerging evidences indicate that lncRNAs are involved in tumour initiation and progression through the regulation of gene expression at the epigenetic, transcriptional, and post-transcriptional levels.6,7 Aberrant lncRNAs exert effects on cancer detection, and circulating lncRNAs have been reported as potential biomarkers for the diagnosis and prognosis of different cancers.8 For example, plasma lncRNA H19 has been identified in GC and may serve as a potential biomarker for GC diagnosis.9 Additionally, different plasma lncRNAs panels can be used as biomarkers for GC detection.10,11 These data indicate that plasma lncRNAs may serve as biomarkers for GC.
Exosomes, one of the extracellular vesicles with a size in the range of 50–150 nm, are secreted by many cells and involved in intercellular communication by transmitting intracellular cargos, such as nucleic acids, lipids, and proteins, including lncRNA.12 Exosomes secreted from tumour cells can be transferred to the circulation, and exosomal RNAs are correlated with the clinicopathological features of cancer.13,14 Furthermore, exosomes can protect RNAs from degradation, and the stability and longevity of RNA molecules are ideal for noninvasive tumour diagnosis.15 Hence, we supposed that GC cells (GCCs) can generate and release exosomes, and the examination of plasma exosomal lncRNAs may provide a convenient and noninvasive method for GC diagnosis and prognosis.
In the present study, we systematically investigated the dysregulated lncRNAs in GCC-derived exosomes according to lncRNA microarray results. Data showed that lnc-SLC2A12-10:1 was significantly upregulated in GCC exosomes and plasma of GC patients. The diagnostic potential of plasma exosomal lnc-SLC2A12-10:1 was further investigated, and its relationship with the clinicopathological features of GC patients was evaluated.
Patients and Methods
Cell Culture
Four human GCC lines (MGC803, BGC823, SGC7901, and AGS) and one normal human gastric (GES-1) cell line were purchased from the Chinese Academy of Sciences Cell Bank of Type Culture Collection. All cell lines were cultured at 37 °C with 5% CO2 in 1640/DMEM (HyClone) containing 10% exosome-depleted FBS supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco). Culture media (CM) were collected, centrifuged at 2000 g for 10 min, followed by another 12,000 g for 10 min at 4 °C, and then stored at −80 °C until exosome extraction.
Patients
Plasma samples from 60 GC patients and 60 age-matched healthy controls were collected from Henan Provincial People’s Hospital between February 2018 and March 2019. All enrolled patients were newly diagnosed GC subjects. The inclusion criteria were as follows: (1) all subjects were diagnosed by gastroscopy or histopathology and (2) did not undergo any preoperative treatment. The exclusion criteria were as follows: (1) combined with other cancer patients and (2) combined with heart, kidney, and liver dysfunction. All clinicopathological data for the GC samples, including age, sex, clinical stage, and histological grade, were obtained from the clinical and pathological records. Moreover, 20 paired GC tissues and their adjacent normal tissues were obtained from resection before other therapies were performed. This study was performed in accordance with the rules of the Declaration of Helsinki of 1975 (revised in 2013) and approved by the Ethics Committee of Henan Provincial People’s Hospital. Informed consent forms were obtained from all subjects before they participated in the study. Plasma samples were separated within 2 h after collection following a two-step centrifugation protocol (3000 g for 10 min at 4 °C, 12,000 g for 10 min at 4 °C) to thoroughly remove cell debris. The separated plasma was stored in RNase-free centrifuge tubes at −80 °C until exosome extraction.
Exosome Isolation and Identification
Plasma and CM were filtered through a 0.22 μm pore polyvinylidene fluoride filter (Millipore). Subsequently, exosomes were isolated using ExoQuick solution or ExoQuick-TC solution (System Biosciences, USA) according to the manufacturer’s protocol. The exosome pellets were resuspended in 50 μL of PBS and stored at −80 °C until further analysis. The concentration and size distribution of the exosomes were determined using nanoparticle tracking analysis (NTA) on a Zeta Potential/Particle Sizer NICOMPTM 380 ZLS analyzer (Santa Barbara, California, USA) following our previous description.16
Transmission Electron Microscopy (TEM)
For TEM, 10 μL of exosome suspension was absorbed onto carbon-coated copper grids (200 mesh) for 1 min. The samples were washed with double-distilled water and negatively stained with 2% uranyl acetate solution for 1 min. After air-drying, the samples were visualized at 87,000× in a Phillips Tecnai TEM at 80 kV.
Western Blot
Briefly, the cells or exosomes were lysed in standard RIPA buffer supplemented with protease and phosphatase inhibitor cocktails (Roche). The amount of proteins was measured with a BCA protein assay kit (Beyotime, China). The protein was solubilized with loading buffer (5×) and heated at 100°C for 10 min. Proteins were separated by SDS-PAGE and then transferred to a 0.2-μm PVDF membrane (Bio- Rad, USA). After blocking with Odyssey Blocking Buffer (Li-COR Biosciences, USA), the membrane was incubated with primary antibody (1:1000) at 4°C overnight, then incubated with IRDye 800CW or 680 secondary antibodies (1:5000, LI-COR Biosciences, USA). The Odyssey Infrared Imaging System was used to visualize targeted protein bands.
lncRNA Microarray Analysis
Exosome pellets from 10 mL of CM were collected for lncRNA microarray assay. Total RNA was extracted using miRNeasy Micro Kit (Qiagen) and quantified with a NanoDrop 2000c spectrophotometer (Thermo Scientific). The quality of RNA was assessed by capillary electrophoresis on an Agilent 2100 Bioanalyzer (Agilent Technologies, CA). The lncRNA microarray analysis was performed by Shanghai Biotechnology Corporation using the Agilent human lncRNA V6 Microarray. The microarray data have been uploaded to the GEO database (GSE148334).
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
The total RNA from tissues/cells was extracted using TRIzol reagent (Invitrogen), and exosomal RNA was extracted using a miRNeasy Micro Kit (QIAGEN) according to the manufacturers’ instructions. The quantity and quality of the RNA were evaluated using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Purified RNA was reversely transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara). Then, qRT-PCR was performed using SYBR Green assays (Takara) on an ABI 7900 system (Applied Biosystems). The reactions were incubated at 95 °C for 10 min, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s. All experiments were conducted in triplicate, and the products were confirmed by melting curve analysis following each reaction. The level of each candidate lncRNA was normalized to that of GAPDH. The primers used for the lnc-SLC2A12-10:1 were (forward) 5′-CACTGAAACCAGCTTGCAT-3′ and (reverse) 5′-TCACTCCCTTGGCTATGAGG-3′. GAPDH: (forward) 5′-CTCTGCTCCTCCTGTTCGAC-3′ and (reverse) 5′-GCGCCCAATACGACCAAATC-3′. The relative expression levels of the target lncRNAs were calculated using the 2−ΔΔCT method.
Serum Tumour Marker Assay
The expression levels of CEA, CA19-9, and CA72-4 were determined according to the analysis requirements of Roche COBAS E602. The CEA, CA19-9, and CA72-4 reference intervals are 0–5.0 ng/mL, 0–35.0 U/mL, and 0–6.9 U/mL, respectively.
Statistical Analysis
All statistical analyses were performed with SPSS 20.0 and GraphPad Prism 7.0 Software. The relationships of the expression level of plasma exosomal lnc-SLC2A12-10:1 with clinicopathological parameters were evaluated by a Chi-square test. Other statistical significance between groups was determined by a Student’s t-test or one-way ANOVA. Receiver operating characteristic (ROC) curve and area under curve (AUC) were used to estimate the diagnostic value of each index for GC. A combined ROC was calculated on the basis of the logistic regression model. P value <0.05 was regarded as statistically significant.
Results
Characterization of Purified Exosomes
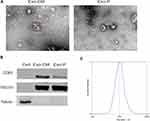
Exosomes are characterized by their typical morphology and conserved size, as well as the presence of specific protein markers. To ensure that exosomes were extracted successfully, we identified the exosomes isolated from the CM (Exo-CM) and plasma (Exo-P) through TEM, particle size analysis, and Western blot. The purified exosomes displayed a typical structure, with a diameter ranging from 80 nm to120 nm (Figure 1A). Conventional exosomal protein markers CD63 and TSG101 were detected in the exosome samples but not in the GCCs (Figure 1B). The expression of CD63 was relatively higher when exosomes were isolated using commercial kit. NTA showed the size distribution of exosomes, which was consistent with the range of exosomes reported in the literature (Figure 1C). The characterization of the exosome purified from cell CM was the same as that from plasma. Hence, these results confirm that exosomes were successfully purified from CM and plasma.
lncRNA Microarray Assay for Identification of GC-Specific Exosomal lncRNA
Exosomal RNA from the CM of four GCCs and GES-1 were purified, and lncRNA microarray assay was conducted for the identification of GC-specific exosomal lncRNAs. After the combined analysis, 199 exosomal lncRNAs were found to be upregulated (fold change of ≥2 and P ≤ 0.05) in CM from GCCs compared with those in GES-1 (Figure 2A). The heatmap showed the different expression levels of the 199 upregulated lncRNAs, and we preferentially selected the top 10 upregulated lncRNAs for further verification (Figure 2B).
Verification of Upregulated Exosomal lncRNAs in GC Specimens and GCCs
To confirm the robustness of the lncRNA microarray assay, we selected the top 10 upregulated lncRNAs for further validation in cell and exosomes from CM and plasma. qRT-PCR showed that the top 10 upregulated lncRNAs increased in GCCs and GCC CM-derived exosome, which was consistent with the microarray results (data not shown). Among these upregulated lncRNAs, one particular lncRNA (lnc-SLC2A12-10:1) was significantly upregulated in the plasma exosomes and tissues of the patients with GC compared with controls (Figure 3). These results suggested that exosomal lnc-SLC2A12-10:1 may be a potential noninvasive biomarker for GC diagnosis.
Expression Pattern and Diagnostic Accuracy of Plasma Exosomal lnc-SLC2A12-10:1 in Patients with GC
To further investigate whether lnc-SLC2A12-10:1 can be used as a biomarker for GC diagnosis, we analyzed the lnc-SLC2A12-10:1 expression levels in 60 patients with GC and 60 healthy controls by qRT-PCR. The exosomal lnc-SLC2A12-10:1 expression level was significantly upregulated in GC patients compared with healthy controls. The ROC curve and AUC were also generated to evaluate the diagnostic accuracy of lnc-SLC2A12-10:1 and compared with the conventional tumour marker (CEA, CA19-9, CA72-4). The AUC value of exosomal lnc-SLC2A12-10:1 was 0.776 (95% confidence interval [CI]: 0.694–0.858, P < 0.001), which was significantly higher than those of CEA, CA 19–9, and CA 72–4, of 0.677 (95% CI: 0.582–0.772, P < 0.001), 0.660 (95% CI: 0.563–0.758, P < 0.01), and 0.633 (95% CI: 0.533–0.732, P < 0.05), respectively (Figure 4A). At the optimal cut-off value, the sensitivity and specificity of exosomal lnc-SLC2A12-10:1 were 78.3% and 75.0%, respectively. The AUC value increased to 0.851 (95% CI 0.780–0.922, P < 0.001) when combined with CEA, CA 19–9, and CA 72–4. The combinative diagnostic capability was better than that of each marker alone (Figure 4B). These results showed that exosomal lnc-SLC2A12-10:1 may be an appropriate diagnostic marker for GC.
Correlation of Plasma Exosomal lnc-SLC2A12-10:1 with Clinicopathological Characteristics
The association between exosomal lnc-SLC2A12-10:1 expression levels and the clinicopathological features of GC patients was also analysed. A total of 60 GC patients were classified into relatively high and low expression groups according to the cut-off value of the ROC curve. Statistical analysis showed that lnc-SLC2A12-10:1 expression levels were positively associated with tumour size (P = 0.035), TNM stage (P = 0.018), lymph node metastasis (P = 0.032), and differentiation (P = 0.017) of GC. Conversely, the correlation of the lnc-SLC2A12-10:1 expression with other clinical features, such as age, gender, CEA, CA19-9, and CA72-4, was not significant (Table 1).
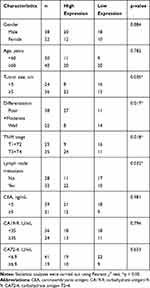 |
Table 1 Correlation Between lnc-SLC2A12-10:1 Expression Levels and Clinicopathologic Features of GC Patients |
Dynamic Monitoring of Plasma Exosomal lnc-SLC2A12-10:1 in GC Patients
To determine whether the expression levels of lnc-SLC2A12-10:1 are correlated with tumour load and revert to a normal state after surgery, we investigated the differences among the lnc-SLC2A12-10:1 levels of 12 paired preoperative/postoperative cases. The lnc-SLC2A12-10:1 expression levels significantly decreased 10 days after surgery (P < 0.05, Figure 5). The results indicated that exosomal lnc-SLC2A12-10:1 may be used for dynamic monitoring during the treatment of GC patients.
 |
Figure 5 lnc-SLC2A12-10:1 expression levels in 12 paired GC patients before and after surgery. |
Discussion
Early diagnosis and prompt treatment can substantially improve the overall survival rate of GC patients.17 At present, the main methods of GC diagnosis include X-ray, gastroscopy, CT, and laboratory examinations. Endoscopy followed by pathological examination is the gold standard for GC, but it is invasive and uncomfortable, leading to a limited use of widespread screening in China.18 Biomarkers in plasma and serum provide a convenient and noninvasive method for tumour diagnosis. However, the current serum tumour biomarkers used for GC diagnosis, such as CEA, CA 19–9, and CA 72–4, exhibit low sensitivity and specificity, particularly at the early stage of cancer.19 As a promising alternative to liquid biopsy, circulating tumour-derived exosomes have gained interest in the fields of noninvasive cancer diagnosis and treatment response monitoring.20
Tumour-derived ncRNAs, including lncRNAs, can be secreted into body fluids, such as blood, urine, milk, and saliva via exosomes.13,21 Exosomal lncRNAs are more stable and concentrated than general lncRNAs in peripheral circulation due to the protection of exosomal membranaceous structures.22 Tumour-derived exosomal RNAs can also accurately reflect the changes in cancer cells during tumour progression.23 Thus, circulating exosomal lncRNAs have emerged as reliable and efficient hallmarks for cancer diagnosis and prognosis monitoring.
In recent years, numerous studies have demonstrated that circulating lncRNAs could function as biomarkers in several cancers, including GC. For instance, serum exosomal lncRNA CRNDE-h can effectively distinguish CRC patients from benign colorectal diseases and NC subjects, with a significantly high AUC value of 0.892, and is significantly correlated with aggressive tumour behavior and poor prognosis.15 A recent study identified three upregulated serum exosomal lncRNAs (PCAT‐1, UBC1, and SNHG16) and designed a three-lncRNA panel, which offers an easy and fast noninvasive approach for the diagnosis and recurrence prediction of bladder cancer.24 In addition, serum exosomal lncRNAs ENSG00000258332.1 and LINC00635 were used for hepatocellular carcinoma diagnosis and prognosis.25 For GC research, serum exosomal lncRNA HOTTIP is typically increased in GC, and exosomal HOTTIP overexpression is an independent prognostic factor in GC patients.26 Early GC-specific exosomal lncRNAs (lncUEGC1) is identified through RNA sequencing and have been proved a convincing biomarker for early-stage GC.27 Consistent with previous studies, our study first identified GC-specific exosomal lncRNA through lncRNA microarray assay. Then, we primitively validated that exosomal lnc-SLC2A12-10:1 was significantly upregulated in the plasma of GC patients and had a certain diagnostic value. Cell CM was selected instead of direct plasma samples because acquiring a considerable amount of total RNA that can be used for microarray is difficult due to the limited exosomes in the plasma, and the exosomal lncRNAs derived from GCCs may be exclusive to GC.
Among the top 10 upregulated lncRNAs, lnc-SLC2A12-10:1 was the uniquely upregulated lncRNA in the plasma-derived exosome of GC patients. Thus, we focused on this special exosomal lncRNA in our subsequent work. We further evaluated the diagnostic performance of exosomal lnc-SLC2A12-10:1 for GC detection. Plasma exosomal lnc-SLC2A12-10:1 exhibited an AUC value of 0.776 in distinguishing GC from healthy volunteers in our cohort of subjects, which was better than those of traditional serum tumour markers (CEA, CA 19–9, and CA 72–4). Furthermore, we found that the combination of CEA, CA 19–9, CA 72–4, and lnc-SLC2A12-10:1 would more accurately discriminate between cases and controls. The AUC of the combination can be increased to 0.851, and the diagnostic efficacy was similar to those of previous studies.27,28 In the present study, the expression level of exosomal lnc-SLC2A12-10:1 was also significantly correlated with tumour size, TNM stage, lymph node metastasis, and differentiation. The exosomal lnc-SLC2A12-10:1 expression level significantly decreased 10 days after surgery compared with before surgery. Despite the relatively small sample size, the results of this study suggested that exosomal lnc-SLC2A12-10:1 may be a useful prognostic indicator.
lnc-SLC2A12-10:1 is located on chromosome 6, with a length of 701 nt. To the best of our knowledge, lnc-SLC2A12-10:1 has not been investigated before. Other studies have demonstrated that lncRNAs are involved in tumour initiation and progression through gene expression regulation.6 Understanding the function of plasma lncRNAs in tumorigenesis would facilitate their clinical application. Our preliminary tests showed that lnc-SLC2A12-10:1 may be a carcinogenic factor that can promote GCC proliferation and invasion in vitro (data not shown).
Taken together, our study suggested that exosomal lnc-SLC2A12-10:1 may be a potential noninvasive biomarker for GC diagnosis and prognosis monitoring. Of course, we recognized that this is a preliminary report on the clinical values of exosomal lnc-SLC2A12-10:1 in GC. Further studies with large-scale clinical samples, dynamic monitoring, and long-term follow up are necessary to support our conclusions. Functional analysis is also required to elucidate the possible regulating mechanism of exosomal lncRNAs in GC progression.
Funding
This study was funded by the Jiangsu Provincial Commission of Health and Family Planning (No. Q2017010), the Jiangsu Provincial Medical Youth Talent (No. QNRC2016781), and the National Natural Science Foundation of China (No. 81802094).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
2. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
4. Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388:2654–2664. doi:10.1016/S0140-6736(16)30354-3
5. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi:10.1016/j.cell.2014.03.008
6. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
7. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887
8. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;9085195.
9. Zhou X, Yin C, Dang Y, et al. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5(1):11516. doi:10.1038/srep11516
10. Liu J, Wang J, Song Y, et al. A panel consisting of three novel circulating lncRNAs, is it a predictive tool for gastric cancer? J Cell Mol Med. 2018;22:3605–3613. doi:10.1111/jcmm.13640
11. Zheng R, Liang J, Lu J, et al. Genome-wide long non-coding RNAs identified a panel of novel plasma biomarkers for gastric cancer diagnosis. Gastric Cancer. 2019;22:731–741. doi:10.1007/s10120-018-00915-7
12. Nabet BY, Qiu Y, Shabason JE, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017;170:352–366. doi:10.1016/j.cell.2017.06.031
13. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi:10.1016/j.ccell.2016.03.004
14. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–1232. doi:10.1016/j.cell.2016.01.043
15. Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi:10.18632/oncotarget.13465
16. Li SB, Zhao Y, Chen WB, et al. Exosomal ephrinA2 derived from serum as a potential biomarker for prostate cancer. J Cancer. 2018;9(15):2659–2665. doi:10.7150/jca.25201
17. Park JY, von Karsa L, Herrero R. Prevention strategies for gastric cancer: a global perspective. Clin Endosc. 2014;47:478–489. doi:10.5946/ce.2014.47.6.478
18. Sawaki K, Kanda M, Kodera Y. Review of recent efforts to discover biomarkers for early detection, monitoring, prognosis, and prediction of treatment responses of patients with gastric cancer. Expert Rev Gastroenterol Hepatol. 2018;12:657–670. doi:10.1080/17474124.2018.1489233
19. Li TT, Liu H, Yu J, et al. Prognostic and predictive blood biomarkers in gastric cancer and the potential application of circulating tumor cells. World J Gastroenterol. 2018;24:2236–2246. doi:10.3748/wjg.v24.i21.2236
20. Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi:10.3410/B3-15
21. Ragusa M, Barbagallo C, Cirnigliaro M, et al. Asymmetric RNA distribution among cells and their secreted exosomes: biomedical meaning and considerations on diagnostic applications. Front Mol Biosci. 2017;4:66. doi:10.3389/fmolb.2017.00066
22. Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi:10.1186/1471-2164-14-319
23. Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. 2016;15:249–256. doi:10.1093/bfgp/elv058
24. Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23:1396–1405. doi:10.1111/jcmm.14042
25. Xu H, Chen Y, Dong X, et al. Serum exosomal long noncoding RNAs and for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27:710–716. doi:10.1158/1055-9965.EPI-17-0770
26. Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17(1):68. doi:10.1186/s12943-018-0817-x
27. Lin LY, Yang L, Zeng Q, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84. doi:10.1186/s12943-018-0834-9
28. Zong W, Feng W, Jiang Y, et al. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin Chem Lab Med. 2019;57:1063–1107. doi:10.1515/cclm-2018-0929
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.