Back to Journals » OncoTargets and Therapy » Volume 12
Piwi-Interacting RNA1037 Enhances Chemoresistance and Motility in Human Oral Squamous Cell Carcinoma Cells
Authors Li G, Wang X, Li C, Hu S, Niu Z, Sun Q, Sun M
Received 3 October 2019
Accepted for publication 13 November 2019
Published 4 December 2019 Volume 2019:12 Pages 10615—10627
DOI https://doi.org/10.2147/OTT.S233322
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Guanghui Li,* Xi Wang,* Chunmei Li,* Shuang Hu, Zhixing Niu, Qiang Sun, Minglei Sun
Department of Stomatology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qiang Sun; Minglei Sun
Department of Stomatology, The First Affiliated Hospital of Zhengzhou University, 1 East Jianshe Road, Zhengzhou, Henan 450052, People’s Republic of China
Tel +86-371 6691 3345
Fax +86-371-6691 3935
Email [email protected]; [email protected]
Background: Piwi-interacting RNAs (piRNAs) are thought to silence transposable genetic elements. However, the functional roles of piRNAs in oral squamous cell carcinoma (OSCC) remain unelucidated. In the present study, we aimed to investigate the role of Piwi-interacting RNA 1037 (piR-1037) in chemoresistance to cisplatin (CDDP)-based chemotherapy and the oncogenic role of piR-1037 in OSCC cells.
Methods: RT-PCR was used to evaluate the levels of piR-1037 and X-linked Inhibitor of apoptosis protein (XIAP) mRNA in OSCC cell lines or tumor xenografts. Transfection of piR-1037 DNA antisense and piR-1037 RNA oligonucleotides was performed to suppress and overexpress piR-1037 in OSCC cells, respectively. A CCK8 assay was used to measure the viability or proliferation of OSCC cells. Apoptosis in OSCC cells and xenografts was determined using a TUNEL assay kit. The activity of caspase-3, caspase-8 and caspase-1 in OSCC cells was measured with colorimetric caspase assay kits. Western blot analysis was conducted to analyze XIAP expression in OSCC cells and xenograft samples. Immunoprecipitation (IP) and RNA pull-down assays were utilized to analyze the piR-1037 - XIAP interaction. Transwell assays were performed to evaluate migration and invasion of OSCC cells.
Results: CDDP treatment upregulated piR-1037 expression in OSCC cells and OSCC xenografts. Suppression of the CDDP-induced upregulation of piR-1037 expression enhanced the sensitivity of OSCC cells to CDDP. piR-1037 promoted protein expression and directly bound XIAP, a key apoptotic inhibitor that is implicated in chemoresistance. The relationship between piR-1037 and XIAP suggested that piR-1037 enhanced OSCC cell chemoresistance to CDDP at least partially through XIAP. Moreover, targeting the basal expression of piR-1037 inhibited cell motility by affecting epithelial–mesenchymal transition (EMT).
Conclusion: piR-1037 enhances the chemoresistance and motility of OSCC cells. piR-1037 promotes chemoresistance by interacting with XIAP and regulates the motility of OSCC cells by driving EMT.
Keywords: piwi-interacting RNA 1037, oral squamous cell carcinoma, cisplatin, apoptosis, X-linked inhibitor of apoptosis protein, motility
Introduction
OSCC is among the most common and aggressive types of head and neck cancers. So far, the mainstay treatment modalities for OSCC are surgery, radiation and chemotherapy. Despite the development of new strategies for OSCC treatment such as gene therapy and therapy targeting epidermal growth factor receptor (EGFR),1–4 the overall prognosis and survival rate of OSCC patients, especially those with an advanced stage of oral cancer, still remain poor. Patients with advanced or metastatic tumor are usually advised chemotherapy. CDDP-based chemotherapy is the most commonly used chemotherapeutic regimen for the treatment of OSCC. CDDP is the first platinum-based drug that is highly potent against cancers and it is the backbone agent in CDDP-based chemotherapy.2 However, chemoresistance constitutes to be a major cause of failure of treatment. Therefore, it is urgent to better understanding the molecular mechanism underlying chemoresistance in patients with OSCC.
piRNAs are a novel group of functional small noncoding RNAs (sncRNAs) that are 24–34 nucleotides in length, characterized by a 3-terminal 2ʹ-O methylation and exclusively associated with piwi superfamily proteins.5–8 piRNAs are able to protect the integrity of the genome by modulating the activity of transposons.9 piRNAs are expressed not only in the germline but also in somatic tissues and exhibit tissue specificity.8 Recent research has proven that piRNAs are implicated in various human cancers, such as lung, gastric, breast, renal, and colorectal cancers.8,10–13 Li et al reported that in a comparison of localized and metastatic clear-cell renal cell carcinoma (ccRCC), piR-1037 expression was significantly upregulated in metastatic ccRCC.14 However, the roles of piR-1037 in cancers, including OSCC, still remain largely unknown.
XIAP belongs to the family of intrinsic inhibitors of apoptosis (IAP) proteins15 and is one of the most critical inhibitors of apoptosis. XIAP plays an essential role in cell survival by blocking the intrinsic and extrinsic apoptotic pathways.16 XIAP contains BIR2 and BIR3 domains that can neutralize caspase-3, caspase-7 and caspase-9, the chief molecules responsible for cell apoptosis.17 Given its inhibitory role in cell apoptosis, XIAP is critical for cancer progression and chemoresistance in cancers, such as lymphoma and oral, lung, breast, renal and liver cancers, and is considered a cancer biomarker.18–24
In the present study, we sought to investigate the roles of piR-1037 in CDDP-based chemoresistance and cell motility in OSCC. We identified that piR-1037 expression was significantly upregulated in response to CDDP chemotherapy in OSCC. Moreover, we observed that suppression of the piR-1037 upregulation induced by CDDP chemotherapy promoted the sensitivity of OSCC cells to CDDP. We then sought to explore the molecular mechanism underlying the contribution of piR-1037 to the chemoresistance of OSCC to CDDP, and we found that piR-1037 increased the protein levels of XIAP and that piR-1037 directly bound the XIAP protein, suggesting that piR-1037 enhanced chemoresistance in OSCC cells in response to CDDP treatment at least partially through XIAP. In addition, targeting the basal expression of piR-1037 in OSCC cells inhibited migration and invasion by driving EMT. Our findings suggest that targeting piR-1037 may be a promising strategy for limiting chemoresistance and metastasis in OSCC.
Methods
Cell Culture
SCC4, SCC9, SCC15 and SCC25 OSCC cells were obtained from American Type Culture Collection (ATCC, USA). UM-SCC6 and UM-SCC-1 were from Sigma Aldrich, USA. HaCat cells were from Thermo Fisher Scientific, Inc. The OSCC cell lines were cultured in Dulbecco’s modified Eagle’s medium/F12 (DME F12, Sigma Aldrich, USA) with 10% fetal bovine serum (FBS), 400 ng/mL of hydrocortisone, and 1× penicillin/streptomycin (Sigma Aldrich, USA). Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 1× penicillin-streptomycin was used for the culture of HaCat cells. The cells were cultured at 37°C, 5% CO2 and 95% humidity in a tissue culture incubator.
CCK8 Assay
Cell viability was determined using CCK8 assay (Dojindo, Japan) according to the manufacturer’s instruction. 100 μL of cell suspension was dispensed in 96-well plates (Corning, USA) at a density of 5000 cells/well. The culture plates were pre-incubated for 24 hrs in a humidified incubator. The cells were cultured for 48 hrs followed by adding 10 μL of CCK8 reagent. The cells were incubated in the presence of CCK-8 for 1 h at 37°C and 5% CO2. The absorbance of optical density (OD) at 450 nm was read in a 800-TS BioTek microplate reader.
RNA Isolation and Real-Time PCR
miRcute miRNA isolation kit (Tiangen Biotech, China) was used to isolate total RNA including piRNAs. Lysis solution was added to the cells or tissues. The aqueous phase containing total RNA was isolated with chloroform and transferred into an elution column to remove protein. The RNAs were eluted in RNAse-free water, quantified and stored at −80 °C. For quantitative analysis of piR1037 expression, 1 ng of total RNA was reverse transcribed using ABI high capacity cDNA archive kit. Real-time PCR was subsequently conducted using a Qiagen miScript SYBR green PCR Kit. The specific primer for piR-1037 was 5ʹ- ACCAGTGGTAGCTACCTGTTATCCATG-3ʹ. The levels of piR-1037 were normalized by Hs_SNORD61_11 miScript primer assay from Qiagen, USA. The PCR amplification program was as follows: 95°C for 5 mins for 1 cycle, 40 cycles of 95°C for 30 s, 60°C for 20 s, and 72°C for 20 s, followed by a final extension at 72°C for 7 mins. For the quantitative assessment of XIAP mRNA expression, total RNA from the cells was extracted using Trizol (Invitrogen, USA). First-strand cDNA was synthesized from 1 μg of RNA using the RevertAid First Strand cDNA synthesis kit from Thermo Fisher Scientific, USA. The PCR amplification program was as follows: 95°C for 10 mins, followed by 32 cycles of 95°C for 1 min, 55°C for 30 s, and 72°C for 45 s, and 1 cycle of 72°C for 5 mins. Primers used for PCR were as follows: forward 5ʹ – GAGAAGATGACTTTTAACAGTTTTGA-3ʹ and reverse 5ʹ- TTTTTTGCT TGAAAGTAATGACTGTGT-3ʹ for XIAP; forward 5ʹ - GTCAGTGGTGGACCTGACCT-3ʹ and reverse 5ʹ- TGAGCTTGACAAAGTGGTCG −3ʹ for GAPDH. PCR was conducted using an ABI PRISM 7500 (Applied Biosystems). The PCR data were analyzed with the 2−ΔΔCt method.
Western Blot
Cells were lysed in lysis buffer containing protease inhibitors (Cell Signaling, USA). BCA protein assay reagent (Thermo Fisher Scientific, USA) was used to determine the concentration of the isolated proteins. 25 μg of the protein was fractioned by SDS-PAGE and afterwards transferred to a PVDF membrane (EMD Millipore, USA). The membrane was then incubated with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 1 h. The membrane was washed once with TBST, followed by incubation with primary antibodies against MDR1 (Novus Biologicals, USA) (1:1000), α-enolase (Boster, China) (1:1000), XIAP (Abcam, USA) (1 μg/mL), cleaved caspase-3 (R&D systems, USA) (0,5 μg/mL), E-Cadherin (1:500), N-Cadherin (1:1000) (Abcam, USA), and β-actin (1:10,000) (Sigma Aldrich, USA). The membrane was then washed three times for 5 mins with TBST on a shaker and incubated with HRP-conjugated goat anti-mouse (1:10,000) or goat anti-rabbit (1:10,000) IgG secondary antibodies (Abcam, USA) and visualized via Immobilon Western Chemiluminescent HRP substrate (EMD Millipore, USA). Densitometry of blots was analyzed using Image J.
Transfection
Complementary DNA oligonucleotides of piR-1037 (piR-1037 antisense) synthesized by Bio Basic, USA, were utilized to target piR-1037. The sequence was 5ʹ-ACCAGTGGTAGCTACCTGTTATCCATGCT-3ʹ. The sequence for scrambled control is 5ʹ- GTACTCTTCGCAGCTGGCAATCGTTCATA-3ʹ. Cell transfection was performed using FuGENE HD transfection reagent (Promega, USA) with 500 nM of piR-1037 antisense or scrambled control and 5 μL of transfection reagent per well in a six-well plate for SCC4 and SCC9 cells. To overexpress piR-1037 in OSCC cells, transfection of OSCC cells with piRNA-1037 RNA oligonucleotides synthesized from BioSynthesis, USA, was conducted using Mirus TansIT-Oligo reagent (Mirus Bio, USA) according to the manufacturer’s instructions with 500 nM of piR-1037 oligonucleotides or its control and 6 μL of transfection reagent. Transfection efficiency was confirmed at 48 h after transfection.
Immunoprecipitation (IP) and Pull-Down Assays
For IP using anti-XIAP antibody, anti-XIAP antibody (Abcam, USA) was used and rabbit IgG served as a negative control. 1 mg of antibody was incubated with 4 mL of Dynabeads protein G (Thermo Fisher Scientific, USA) for 3–4 hrs at 4°C. OSCC cells were washed with cold phosphate-buffered saline (PBS) and lysed with IP lysis buffer (Thermo Fisher Scientific, USA). The protein extracts from OSCC cells pre-cleared using the beads were incubated with the beads saturated with anti-XIAP antibody at 4°C overnight. The beads were then washed with lysis buffer twice. Total RNA was extracted from the beads using miRcute miRNA isolation kit. piR-1037 amplification was performed according to methods described in real-time PCR section. The beads were boiled and the supernatants were used for Western blot to detect XIAP in the immunoprecipitates. For pull-down analysis using piR-1037, biotin-labeled RNA oligonucleotides were synthesized by BioSynthesis, USA. 1 mg of Biotin-labeled RNA was incubated with protein extracts isolated from 5 × 106 OSCC cells using IP lysis buffer overnight at 4°C with rotation. Fifty percent of Streptavidin beads (5 mL) (Sigma Aldrich, USA) were used to couple biotin-labeled RNA. After incubation, the beads were washed five times with lysis buffer. SDS protein loading buffer (Bio Rad, USA) was then used to denature the samples. The proteins in the samples were then analyzed by SDS-PAGE and Western blot using anti-XIAP antibody.
Xenograft
The establishment of the tumor xenografts mouse model was conducted according to the protocols approved (No. 2018036; date: Feb 1, 2018) by the Ethics Committee of Zhengzhou University, Zhengzhou, Henan, China. All methods were performed in accordance with the relevant guidelines and regulations in this approval. Mice were maintained at constant humidity and room temperature (25°C). SCC4 and SCC9 cells (2×106 cells, viability >%95) were 1:1 mixed with Matrigel (BD Biosciences, USA) in 150 μL of serum-free medium and injected subcutaneously into the right dorsal-lateral side of 6-week-old female nude mice. Length, width and height of the tumors were measured twice a week using a dial caliper (Mitutoyo, Japan) and tumor sizes were calculated by using the formula: 0.5236 × length × width × height. An i.p. injection of CDDP (1 mg/kg/week) (Sigma Aldrich, USA) was given on day 1, followed by an intratumoral injection of a scramble or piR-1037 antisense DNA oligos at 5 μg/50 μL (formulated with Medibena Lancefection siRNA Delivery Agent) on days 4, 6, 8, 10, 12, 14, 16, and 18, respectively. A total of six mice were used for each group. On day 20, mice were euthanized and the tumors were harvested for further study.
Migration and Invasion Assays
Invasion and migration assays were performed using the CytoSelect 24-well Trans-well system (Cell Biolabs, USA) according to the manufacturer’s protocols with slight modification. A total of 1 × 105 of OSCC cells were suspended in 300 μL of serum-free medium supplemented with 0.1% BSA and added to the inserts (pore size: 8 μm). Inserts pre-coated with Matrigel (50 μL/well, Sigma Aldrich, USA) were used for invasion assay. The lower chamber was filled with 500 μL of medium supplemented with 5% FBS. After 12 hrs of incubation, the medium in the inserts was carefully aspirated and non-migratory cells were removed from the interior of the inserts with a cotton swab. The inserts were then stained with a cell stain solution for 15 mins at room temperature (RT) and examined using an Olympus CX31 microscope. Afterward, the inserts were incubated with an extraction solution for 10 mins at RT. 100 μL of extraction solution from each insert was transferred into a 96-well microplate (BD Biosciences, USA) and OD at 560 nm was measured in a 800-TS BioTek microplate reader.
Statistical Analysis
Data were presented as mean ± SD. The difference between two groups was assessed by unpaired student’s t-test. One-way ANOVA was used to analyze the difference among multiple groups. P <0.05 was considered to be statistically significant.
Results
CDDP-Based Chemotherapy Induced the Upregulation of piR-1037 Expression in OSCC Cells
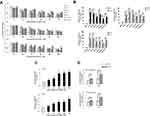
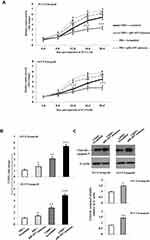
CDDP-based chemotherapy is the combination of CDDP and a chemotherapeutic agent such as 5-FU or paclitaxel (taxol). We first examined the responses of OSCC cell lines to CDDP, 5-FU (Dalian Meilun Biotech, China) or taxol (Bristol-Myers Squibb, USA) by measuring the cell viability of HaCat cells and SCC4, SCC9, SCC15, SCC25, UM-SCC1 and UM-SCC6 OSCC cells treated with different doses of CDDP, 5-FU or taxol. As shown in Figure 1A, at concentrations of 10 μM for CDDP, 5 μM for 5-FU and 50 nM for taxol, the drugs reduced the viability of the six OSCC cell lines by nearly 50%, but there were still a significant number of control HaCat cells that remained alive, which was ideal and important for the role of HaCat cells as a negative control in examining the levels of piR-1037 in OSCC cells. To investigate whether piR-1037 is involved in chemoresistance, we examined the correlations between the levels of piR-1037 and chemotherapy with a fixed dose of CDDP (10 μM), 5-FU (5 μM) or taxol (50 nM) in OSCC cell lines based on the optimization of drug doses, including IC50 determination. We analyzed the changes in the expression levels of piR-1037 in response to the chemotherapeutic agents. We found that CDDP, 5-FU and taxol significantly upregulated piR-1037 expression in the SCC4, SCC9, SCC15, SCC25, UM-SCC1 and UM-SCC6 OSCC cell lines (one-way ANOVA analysis: *p <0.05; **p <0.01) but not in HaCat cells (Figure 1B) (p > 0.05), indicating that piR-1037 expression was correlated with CDDP-based chemotherapy since all the chemotherapeutic agents used in this study could upregulate piR-1037 levels in OSCC cells. Additionally, as shown in Figure 1C, CDDP upregulated piR-1037 expression in a dose-dependent manner in SCC4 and SCC9 cells (one-way ANOVA analysis: *p <0.05; **p <0.01; ***p <0.001). Based on the backbone role of CDDP in CDDP-based chemotherapy, we then used CDDP as a representative agent in the rest of our studies. To further substantiate these findings in vivo, we evaluated the levels of piR-1037 in OSCC xenograft tumors derived from SCC4 and SCC9 cells in xenograft mouse models. The tumors were harvested at 7 days and 20 days post CDDP treatment. We found that the levels of piR-1037 were significantly elevated in the SCC4 and SCC9 tumors at these two time points. Higher levels of piR-1037 were observed in the tumors from the mice that received chemotherapy for 20 days than in those from the mice treated for 7 days (Figure 1D) (one-way ANOVA analysis: *p <0.05), suggesting that the expression of piR-1037 could be enhanced by CDDP therapy in vivo.
Inhibition of piR-1037 in OSCC Cells Promoted OSCC Cell Sensitivity to CDDP
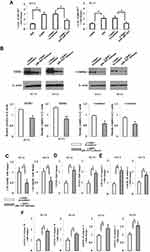
Since we found a correlation between the levels of piR-1037 and CDDP-based chemotherapy in OSCC cells, we then sought to investigate the biological functions of piR-1037 in the chemoresistance of OSCC cells. We inhibited the upregulation of piR-1037 expression induced by CDDP chemotherapy by using piR-1037 antisense DNA oligonucleotides in SCC4 and SCC9 cells. The knockdown efficiency was confirmed as shown in Figure 2A (one-way ANOVA analysis: *p <0.05). We found that piR-1037 antisense oligonucleotides could dramatically inhibit the expression of the chemoresistance biomarkers MDR1 and α-enolase (Figure 2B; Supplementary Figure S1) (unpaired student’s t-test: *p <0.05; **p<0.01) in SCC4 and SCC9 cells treated with CDDP. The results of a CCK8 assay, as indicated in Figure 2C, showed that suppressing piR-1037 significantly reduced the cell viability of SCC4 and SCC9 cells treated with CDDP (unpaired student’s t-test: *p <0.05). As shown in Figure 2D, targeting piR-1037 remarkably enhanced cell apoptosis, which was detected using a TUNEL-based colorimetric assay (unpaired student’s t-test: *p <0.05). These results were further substantiated with evidence of increased caspase-3 activity in cells transfected with piR-1037 antisense oligonucleotides (Figure 2E) (unpaired student’s t-test: *p <0.05). These findings suggest that targeting piR-1037 increases the sensitivity of the OSCC cells to CDDP treatment, as evidenced by an increase in cell apoptosis and downregulation of chemoresistance biomarker expression. Surprisingly, we found that targeting piR-1037 could increase the levels of activity of both caspase-8 and caspase-1 (Figure 2F) (unpaired student’s t-test: *p <0.05), suggesting that silencing the CDDP-induced upregulation of piR-1037 expression could trigger redundant cell death pathways in OSCC cells treated with CDDP chemotherapy.
Suppression of piR-1037 Promoted CDDP-Induced Apoptosis in Xenograft Mouse Models
We then sought to determine whether piR-1037 is correlated with CDDP chemosensitivity in animal models. A tumor xenograft model was established to determine whether targeting piR-1037 promotes cell apoptosis in tumors derived from the SCC4 and SCC9 cell lines in nude mice. We first confirmed the suppressive effect of piRNA-1037 antisense oligonucleotides on the expression of piRNA-1037 in tumor xenografts on days 4, 8, 12, 16 and 20 (Supplement Figure S2). We found that the size of tumors in mice treated with CDDP and piR-1037 antisense oligonucleotides was dramatically smaller than that in mice treated with CDDP and a scrambled control (Figure 3A) (one-way ANOVA analysis: *p <0.05; **p <0.01; ***p <0.001), suggesting that piR-1037 is involved in the growth of tumor treated with a chemotherapeutic agent. Moreover, apoptosis and the expression of cleaved caspase-3 were significantly increased in tumor xenografts from the mice treated with the combination of CDDP and piR1037 antisense oligonucleotides compared with those from the mice administered CDDP and the scrambled control (Figure 3B and C; Supplementary Figure S3) (one-way ANOVA analysis or unpaired student’s t-test: *p <0.05; **p <0.01; ***p <0.001).
Binding of piR-1037 to XIAP in OSCC Cells
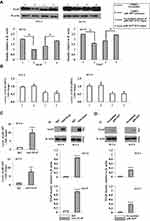
Next, we investigated the mechanisms underlying the role of piR-1037 in the regulation of chemosensitivity in OSCC cells. Since piR-1037 can suppress apoptosis in OSCC cells treated with chemotherapy and XIAP is a pivotal inhibitor of apoptosis-related proteins in tumor cells that can neutralize caspases and a critical protein in cancer chemoresistance, we hypothesized that piR-1037 may be a regulator of XIAP. To determine whether the role of piR-1037 in chemoresistance in OSCC cells is correlated with XIAP, we examined the levels of XIAP in OSCC cells treated with CDDP in combination with piR-1037 antisense oligonucleotides or forced expression of piR-1037 only. We found that the protein levels of XIAP were dramatically elevated in the cells with forced overexpression of piR-1037 and that piR-1037 antisense oligonucleotides exerted the opposite effect on cells treated with CDDP (Figure 4A, Supplementary Figure S4) (one-way ANOVA analysis: *p <0.05). Neither suppression nor overexpression of piR-1037 significantly altered the mRNA levels of XIAP, which are usually controlled at the transcriptional level (Figure 4B) (p > 0.05), suggesting that piR-1037 could regulate the expression of XIAP at the posttranscriptional level by maintaining XIAP mRNA stability, enhancing XIAP translation or inhibiting protein degradation. On the basis of these findings, we then considered whether piR-1037 directly binds XIAP. An IP assay was then performed to detect any direct interaction between piR-1037 and XIAP. As shown in Figure 4C and Supplementary Figure S5, piR-1037 was detected in XIAP IP products (unpaired student’s t-test: **p <0.01; ***p <0.001). The results were further confirmed by a pull-down assay with biotinylated piR-1037 RNA oligos in OSCC cells treated with CDDP (Figure 4D; Supplementary Figure S5) (unpaired student’s t-test: ***p <0.001).
piR-1037 Enhanced the Motility of OSCC Cells
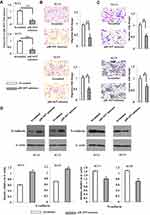
To evaluate the biological function of the basal levels of piR-1037 in OSCC cells, the expression of piR-1037 was silenced by piR-1037 antisense oligonucleotides (Figure 5A) (unpaired student’s t-test: **p <0.01). Cell motility and the expression of the EMT biomarkers E-cadherin and N-cadherin were assessed. We found that targeting piR-1037 dramatically inhibited cell migration (Figure 5B) and invasion (Figure 5C) (unpaired student’s t-test: *p <0.05; **p <0.01) and the expression of E-cadherin, the epithelial marker, was upregulated, whereas the levels of the mesenchymal marker N-cadherin were reduced in the SCC4 and SCC9 cell lines (Figure 5D; Supplementary Figure S6) (unpaired student’s t-test: *p <0.05). These findings suggest that in addition to its association with chemoresistance, piR-1037 can also affect the motility of OSCC cells.
Discussion
piRNAs were originally identified to modulate the activity of transposons and participate in the regulation of the expression of protein-coding genes and chromatin modification.25–28 Recent studies have revealed that aberrant expression of piRNAs in several types of cancers is correlated with cancer phenotypes.10,29,30 In this study, we examined the effects of piR-1037 on chemoresistance to CDDP and its oncogenic role in OSCC cells. Our findings suggest that piR-1037 expression levels are closely correlated with chemoresistance to CDDP chemotherapy in OSCC and that piR-1037 can promote chemoresistance in OSCC cells, at least partially through the XIAP-caspase axis. In addition, we found that the basal expression of piR-1037 exerted an oncogenic effect on OSCC cells.
Our findings showed that the upregulation of piR-1037 expression could enhance chemoresistance by inhibiting apoptosis induced by CDDP chemotherapy and the expression of chemoresistance biomarkers in OSCC cells. The cleaved form of caspase-3 has been widely demonstrated to be a critical protease in the apoptotic signaling pathway and a checkpoint indicating the initiation of cell apoptosis.31,32 Previous studies have proven that activated caspase-8 is essential for the activation of caspase-3. However, caspase-3 can be activated by activated caspase-1 in the absence of caspase-8.33 In the present study, it was surprising that targeting piR-1037 could increase the levels of both cleaved caspase-1 and cleaved caspase-8, suggesting that the suppression of the CDDP-induced upregulation of piR-1037 expression could trigger redundant cell death processes. It is likely that the suppression of both caspase-8- and caspase-1-dependent death signaling pathways by piR-1037 may be a strategy to allow OSCC cells to cope with the inhibitory effect of CDDP on cell proliferation.
Since piR-1037 contributes to chemoresistance in OSCC cells treated with CDDP chemotherapy by inhibiting apoptosis, suppressing the upregulation of piR-1037 expression induced by CDDP might be a therapeutic strategy for the treatment of OSCC. Here, we investigated the molecular mechanism underlying piR-1037-mediated chemoresistance in OSCC cells. XIAP has been identified as the most potent cell apoptosis-suppressing member of the IAP family.34 It was observed that the protein levels of XIAP but not the mRNA levels were positively correlated with the levels of piR-1037 in OSCC cells treated with CDDP. We then sought to investigate whether piR-1037 directly modulates the function of XIAP by interacting with XIAP in OSCC cells treated with CDDP. Our results for IP and pull-down assays showed that piR-1037 could directly bind XIAP. The direct interaction of piR-1037 with XIAP might contribute to regulating the function of XIAP at the posttranscriptional level, such as protecting XIAP from mRNA decay or protein degradation, promoting the activity of XIAP or enhancing the binding of XIAP to caspases. Further studies are needed to determine the exact molecular mechanism by which piR-1037 modulates the function of XIAP in OSCC cells.
In addition to observing the correlation of piR-1037 with chemoresistance, we also sought to investigate whether piR-1037 exerts an effect on the motility of OSCC cells. The effects of basal levels of piR-1037 on the migration and invasion of OSCC cells were assessed. We demonstrated that silencing the basal expression of piR-1037 with antisense oligonucleotides in OSCC cells could effectively hamper the migration and invasion of OSCC cells, indicating that piR-1037 might also play a role in regulating OSCC metastasis. EMT is a major mechanism of tumor metastasis. Tumor cells undergoing EMT exhibit specific cell phenotypes, such as upregulation of the expression of mesenchymal markers and downregulation of the expression of epithelial markers.35 We found that piR-1037 antisense oligonucleotides could upregulate the expression of E-cadherin, an epithelial marker, and downregulate the expression of the mesenchymal marker N-cadherin in OSCC cells. Therefore, piR-1037 may contribute to OSCC metastasis in addition to its effect on chemoresistance.
Conclusions
This is the first report showing that piR-1037 is associated with chemoresistance and motility in OSCC cells. piR-1037 enhances chemoresistance by interacting with XIAP and regulates the motility of OSCC cells by driving EMT.
Abbreviations
OSCC, oral squamous cell carcinoma; XIAP, X-linked Inhibitor of Apoptosis Protein; IP, Immunoprecipitation; EMT, epithelial–mesenchymal transition; EGFR, epidermal growth factor receptor; sncRNAs, small noncoding RNAs; ccRCC, clear-cell renal cell carcinoma; IAP, inhibitors of apoptosis proteins; FBS, fetal bovine serum; OD, optical density; PBS, phosphate-buffered saline.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Irimie AI, Braicu C, Pileczki V, et al. Knocking down of p53 triggers apoptosis and autophagy, concomitantly with inhibition of migration on SSC-4 oral squamous carcinoma cells. Mol Cell Biochem. 2016;419(1–2):75–82. doi:10.1007/s11010-016-2751-9
2. Georges P, Rajagopalan K, Leon C. Chemotherapy advances in locally advanced head and neck cancer. World J Clin Oncol. 2014;5:966–972. doi:10.5306/wjco.v5.i5.966
3. Ribeiro FA, Noguti J, Oshima CT, et al. Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: a promising approach. Anticancer Res. 2014;34(4):1547–1552.
4. Cripps C, Winquist E, Devries MC, et al. Epidermal growth factor receptor targeted therapy in stages III and IV head and neck cancer. Curr Oncol. 2010;17(3):37–48. doi:10.3747/co.v17i3.520
5. Grivna ST, Beyret E, Wang Z, et al. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi:10.1101/gad.1434406
6. Girard A, Sachidanandam R, Hannon GJ, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi:10.1038/nature04917
7. Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi:10.1126/science.1130164
8. Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5:10423. doi:10.1038/srep10423
9. Siomi MC, Sato K, Pezic D, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi:10.1038/nrm3089
10. Yan H, Wu QL, Sun CY, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29(1):196–206. doi:10.1038/leu.2014.135
11. Muller S, Raulefs S, Bruns P, et al. Next- generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi:10.1186/s12943-015-0358-5
12. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi:10.1038/nrg3074
13. Yin J, Jiang XY, Qi W, et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017;108(9):1746–1756. doi:10.1111/cas.13300
14. Li YP, Wu XW, Gao HL, et al. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med. 2015;21:381–388. doi:10.2119/molmed.2014.00203
15. Holcik M, Gibson H, Korneluk RG. XIAP: apoptotic brake and promising therapeutic target. Apoptosis. 2001;6(4):253–261. doi:10.1023/A:1011379307472
16. Pandey M, Shrivastava M, Mishra S, et al. Role of X-linked inhibitor of apoptosis protein as cancer biomarker. J Clin Diagn Res. 2017;11(12):XE01–XE05.
17. Obexer P, Ausserlechner MJ. X-linked inhibitor of apoptosis protein–a critical death resistance regulator and therapeutic target for personalized cancer therapy. Front Oncol. 2014;4:197. doi:10.3389/fonc.2014.00197
18. Lee YJ, Park BS, Park HR, et al. XIAP inhibitor embelin induces autophagic and apoptotic cell death in human oral squamous cell carcinoma cells. Environ Toxicol. 2017;32(11):2371–2378. doi:10.1002/tox.22450
19. Mandal M, Swan EA, Jasser SA, et al. XIAP can mediate anoikis-resistance in oral squamous cell carcinoma. Cancer Res. 2014;65(9):629.
20. Hofmann HS, Simm A, Hammer A. Expression of inhibitors of apoptosis (IAP) proteins in non-small cell human lung cancer. J Cancer Res Clin Oncol. 2002;128(10):554–560. doi:10.1007/s00432-002-0364-z
21. Ramp U, Krieg T, Caliskan E, et al. XIAP expression is an independent prognostic marker in clear-cell renal carcinomas. Hum Pathol. 2004;35(8):1022–1028. doi:10.1016/j.humpath.2004.03.011
22. Yan Y, Mahotka C, Heikaus S, et al. Disturbed balance of expression between XIAP and Smac/DIABLO during tumour progression in renal cell carcinomas. Br J Cancer. 2004;91(7):1349–1357. doi:10.1038/sj.bjc.6602127
23. Huang X, Wang XN, Yuan XD, et al. XIAP facilitates breast and colon carcinoma growth via promotion of p62 depletion through ubiquitination-dependent proteasomal degradation. Oncogene. 2019;38(9):1448–1460. doi:10.1038/s41388-018-0513-8
24. Shang MJ, Hong DF, Hu ZM, et al. Cisplatin induces apoptosis of hepatocellular carcinoma LM3 cells via down-regulation of XIAP. Eur Rev Med Pharmacol Sci. 2018;22(2):382–387. doi:10.26355/eurrev_201801_14183
25. Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–359. doi:10.1038/nature12987
26. Esposito T, Magliocca S, Formicola D, et al. piR-015520 belongs to Piwi-associated RNAs regulates expression of the human mela- tonin receptor 1A gene. PLoS One. 2011;6:e22727. doi:10.1371/journal.pone.0022727
27. Rajasethupathy P, Antonov I, Sheridan R, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi:10.1016/j.cell.2012.02.057
28. Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356:561–567. doi:10.1016/j.canlet.2014.10.004
29. Peng L, Song L, Liu C, et al. piR-55490 inhibits the growth of lung carci- noma by suppressing mTOR signaling. Tumor Biol. 2016;37:2749–2756.
30. Law PT, Qin H, Ching AK, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi:10.1016/j.jhep.2013.01.032
31. Lavrik IN, Golks A, Krammer PH. Caspases: pharmacological manipulation of cell death. J Clin Invest. 2005;115:2665–2672. doi:10.1172/JCI26252
32. Jessel R, Haertel S, Socaciu C, et al. Kinetics of apoptotic markers in exogenously induced apoptosis of EL4 cells. J Cell Mol Med. 2002;6:82–92. doi:10.1111/jcmm.2002.6.issue-1
33. Sagulenko V, Vitak N, Vajjhala PR, et al. Caspase-1 is an apical caspase leading to Caspase-3 cleavage in the AIM2 inflammasome response, independent of Caspase-8. J Mol Biol. 2018;430(2):238–247. doi:10.1016/j.jmb.2017.10.028
34. Tong QS, Zheng LD, Wang L, et al. Downregulation of XIAP expression induces apoptosis and enhances chemotherapeutic sensitivity in human gastric cancer cells. Cancer Gene Ther. 2005;12(5):509–514. doi:10.1038/sj.cgt.7700813
35. Peinado H, Cano A. New potential therapeutic targets to combat epi- thelial tumor invasion. Clin Transl Oncol. 2006;8:851–857. doi:10.1007/s12094-006-0148-z
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.