Back to Journals » Infection and Drug Resistance » Volume 14
Phenotypic and Genotypic Characteristics of Biofilm Formation in Clinical Isolates of Acinetobacter baumannii
Authors Li Z , Ding Z, Liu Y, Jin X, Xie J, Li T, Zeng Z, Wang Z, Liu J
Received 10 March 2021
Accepted for publication 17 June 2021
Published 7 July 2021 Volume 2021:14 Pages 2613—2624
DOI https://doi.org/10.2147/IDR.S310081
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zhaoyinqian Li,* Zixuan Ding,* Yao Liu, Xinrui Jin, Jingling Xie, Tingting Li, Zhangrui Zeng, Zhibin Wang, Jinbo Liu
Department of Laboratory Medicine, the Affiliated Hospital of Southwest Medical University, Luzhou, 646000, Sichuan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinbo Liu
Department of Laboratory Medical, Affiliated Hospital of Southwest Medical University, 25 Taiping Street, Luzhou, 646000, Sichuan, People’s Republic of China
Tel/Fax +86 0830 3165730
Email [email protected]
Background: Acinetobacter baumannii is an important pathogen in clinical infections, and biofilm formation is an effective way for A. baumannii to survive under external pressures. In this study, the aims were to examine the antimicrobial resistance, biofilm formation, and biofilm-specific resistance in clinical isolates of A. baumannii.
Materials and Methods: A total of 104 clinical A. baumannii isolates were collected from a large teaching hospital in Southwest China. The antibiotics susceptibilities were tested, and biofilm-forming ability was evaluated by crystal violet staining by confocal laser scanning microscopy (CLSM). Minimum inhibitory concentration (MIC), minimum bactericidal concentration (MBC), minimum biofilm inhibitory concentration (MBIC), and minimum biofilm eradication concentration (MBEC) of ciprofloxacin, meropenem, and ceftazidime were tested on selected strains by broth microdilution method. Biofilm-associated genes were detected by polymerase chain reaction (PCR), and expression of genes at planktonic stage and biofilm stage were analyzed by real-time reverse transcription PCR (RT-PCR).
Results: Multidrug-resistant (MDR) isolates accounted for 65.4%, but no strain was resistant to tigecycline and polymyxin B. Moreover, non-MDR strains tended to form stronger biofilms than MDR strains, and a negative correlation between biofilm-forming ability and resistance profiles to each of tested antimicrobials were observed. The MBECs and MBICs of ciprofloxacin, ceftazidime, and meropenem were evidently increased compared with MICs and MBCs among all tested strains. Additionally, the biofilm formation ability of the csuD-positive strains was stronger than that of the csuD-negative strains. The strains in MDR group had higher carrying rate of csuA and csuD genes than non-MDR group, while non-MDR strains possessed more ompA gene than MDR group. Finally, abaI gene was significantly up-regulated after biofilm formation.
Conclusion: These results revealed valuable data for the negative correlation between antimicrobial resistance and biofilm formation, as well as phenotypic and genotypic characteristics of biofilm formation in A. baumannii.
Keywords: Acinetobacter baumannii, biofilm formation, antimicrobial resistance, gene expression
Introduction
Acinetobacter baumannii is one of the most common nosocomial pathogen that causes several severe infections in critically ill patients, and a great number of multidrug-resistant (MDR) strains attract the attention of the whole world.1 The formation of biofilm contributes to A. baumannii adhere to various biological and non-biological surfaces, such as vascular catheters, cerebrospinal fluid shunts, and other medical devices.2 Biofilm is defining composed of extracellular polymeric substances (EPS) that surrounding microorganisms, which mediates microbial adhesion and makes a lifestyle that is totally different from the planktonic state.3 Indeed, biofilm formation presents a significantly higher antimicrobial resistance than planktonic cells, resulting in harder eradication and easier recurrence in biofilm-associated infections.4 Biofilm formation is a complex process and regulated by a variety of factors. Taking A. baumannii for example, biofilm associated protein (Bap) is involved in the initial adhering of biofilm, promoting the maturation of biofilm and maintaining the structure of mature biofilm.5,6 Moreover, synthesis and assembly of pili are encoded by csuA/BABCDE gene cluster to induce the initial adhesion process of biofilm.7 The BfmR-BfmS regulatory system senses the changes of external signals then affects the synthesis and secretion of pili, and induces the biofilm formation by regulating the csuA/BABCDE gene cluster as well.8,9 The pgaABCD locus encodes the production of poly-beta-1-6-N-acetylglucosamine (PNAG), which is not only an important virulence factor but also essential for biofilm formation.10 The self-induced synthetase gene abaI is involved in the quorum sensing (QS) system and plays an important role in later stages of biofilm maturation.11,12 Outer membrane protein A (OmpA) is a well-known virulence factor that contributes to adhesion, invasion, and biofilm formation of A. baumannii.13 On the other hand, the latest research has reported that biofilm growth-associated repressor (BigR) directly or indirectly regulates the expression of genes related to epithelial cell adhesion and biofilm formation.14 Several biofilm-associated genes affect the antimicrobial susceptibility, indicating a relationship between biofilm formation and antimicrobial resistance.11
However, it is still unclear whether there is a quantitative correlation between biofilm formation and antimicrobial resistance. The aims of this study were to analyze the antimicrobial susceptibility, biofilm formation, biofilm-specific resistance, and biofilm-associated gene carrying rate of 104 clinical isolates of A. baumannii. Furthermore, the phenotype-genotype correlation between planktonic and biofilm stage was also evaluated.
Materials and Methods
Identification and Antimicrobial Susceptibility Testing (AST)
A total of 104 non-repetitive A. baumannii strains were isolated from January 2020 to April 2021 in the Affiliated Hospital of Southwest Medical University, and ATCC19606 was used as the reference strain in this study. Identification and antimicrobial resistance analysis were performed by the MicroScan Walk-Away 96 Plus system (Siemens, Germany), and results were interpreted according to the standards of the Clinical and Laboratory Standards Institute (CLSI) 2020-M100.15 Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 (purchased from China National Health Inspection Center) were used as quality control. Based on the obtained antimicrobial resistance profiles, the tested strains were classified into multidrug-resistant (MDR) group if ‘the isolates are resistant to three or more antimicrobial classes’,16 and non-MDR group which the isolates are resistant to 0–2 antimicrobial classes.
Biofilm Formation Assay
Biofilm formation ability was determined by crystal violet staining method as described previously,17 with slight modifications. Briefly, the overnight culture of each strain was diluted to final optical density of 600 (OD600) = 0.1, and then 180 μL of Luria-Bertani (LB) broth (Haibo, Qingdao, China) with 1% glucose (Sigma, USA) and 20 μL of bacterial suspensions were inoculated into 96-well polystyrene microtiter plates (Costar #3524, Corning, NY, USA) in triplicate. After 24 h incubation at 37 °C, the plates were gently washed with phosphate-buffered saline (PBS) for three times and stained with 200 μL of 0.1% crystal violet (Solarbio, Beijing, China) for 20 min. After washing with PBS for three times again, the plates were solubilized with 200 μL of 95% ethanol, followed by shaking slowly at room temperature for 20 min. The absorbance was measured at OD570. The cut-off OD (ODc) was defined as three standard deviations (SD) above the mean OD of the negative control (LB broth only). Following were classification criteria: OD ≤ ODC, non-biofilm producers (-); ODC < OD ≤ 2×ODC, weak biofilm producers (+); 2×ODC < OD ≤ 4×ODC, moderate biofilm producers (++); and OD > 4×ODC, strong biofilm producers (+++).18
Determination of Biofilm Using Confocal Laser Scanning Microscopy (CLSM)
Four strains (two strong biofilm producers, one biofilm-negative producer, and ATCC19606) were selected for biofilm visualization by CLSM (Leica, Germany). In brief, strains were cultured in LB broth for 18 h and diluted to 0.5 McFarland standard. One hundred microlitre suspension and 2 mL fresh LB broth were added in the petri dishes (FengRun, Beijing, China). After incubation at 37 °C for 24 h, the petri dishes were washed three times with PBS and fixed with 2 mL of 2.5% glutaraldehyde for 1.5 h. Then, the petri dishes were washed three times with PBS again and 200 μL fluorescein isothiocyanate-concanavalin A (FITC-ConA) (Sigma, Germany) were added and stored at 4 °C for 30 min. Two hundred microlitre propidium iodide (PI) (Sigma, Germany) were added and stored at 4 °C for 15 min after washing three times with PBS. Finally, the dishes were washed with PBS and sealed with 40% glycerol. FITC-conA binds to EPS of biofilm and reflects green fluorescence at the laser wavelength of 488 nm.19 PI binds to bacterial DNA and emits red fluorescence at 543 nm.20 If both lasers appear at the same time, an orange fluorescence will be shown.21
Growth Curve Assay
The growth of eight strong biofilm producers, eight weak biofilm producers, and ATCC19606 were evaluated. For each strain, 20 μL of diluted overnight culture (0.5 McFarland standard) and 180 μL LB broth were added in 96-well plates and incubated at 37 °C for 24 h, and the OD590 was detected by microplate reader every 2 h until 24 h. After biofilm modeling on plates, the OD570 was measured by microplate reader every 4 h until the number of biofilm biomass decreased. All assays were repeated in triplicate.
Minimum Inhibitory Concentration (MIC), Minimum Bactericidal Concentration (MBC), Minimum Biofilm Inhibitory Concentration (MBIC), and Minimum Biofilm Eradication Concentration (MBEC)
Three antimicrobials that are commonly used for the treatment of A. baumannii infections were selected in the determination of MICs/MBCs and MBICs/MBECs. All of these nine strains were originally sensitive to tested antimicrobials. The MICs for ceftazidime, ciprofloxacin, and meropenem were assessed using broth microdilution method in 96-well plates according to the procedures described in CLSI 2020-M100 guideline.15 After MIC determination, 10 μL aliquots from the wells with no observed bacterial growth were smeared on LB agar plates and incubated at 37 °C for 24 h, and the lowest concentration of antimicrobials that making no colony grows on plates was considered as MBC. AST at biofilm stage was performed according to the procedures described previously.22 Biofilm was established in 96-well plates for 24 h, and planktonic cells were removed by sterile deionized water. Each well was treated with 200 μL of LB broth containing serially diluted antimicrobials. The plates were incubated for 24 h at 37 °C. The minimum antimicrobial concentration at which OD600 < 0.1 was determined as the MBIC.
Following the MBIC examination, the plates were washed with sterile deionized water to remove planktonic cells and antibiotics. Two hundred microlitre of LB broth was added to each well, and the plates were incubated at 37 °C for 24 h. The lowest concentration at which OD600 < 0.1 was considered as the MBEC. All experiments were performed in triplicate.
Detection of Biofilm-Associated Genes by Polymerase Chain Reaction (PCR) and Real-Time Reverse Transcription PCR (RT-PCR)
The DNA template was prepared by boiling bacteria.23 The biofilm-associated genes of all 104 clinical strains were detected by PCR, including bap, ompA, bigR, bfmS, bfmR, csuA, csuB, csuC, csuD, csuE, csuAB, pgaD, and abaI (Table S1). The procedures were followed by previous study with slight modifications.24 Part of nucleotide sequences of PCR products were analyzed by Beijing Qingke Biotechnology Co. Ltd. The results were compared with sequences that retrieved from GenBank database by the BLAST online tool at www.ncbi.nlm.nih.gov. Moreover, eight strong biofilm producers were selected to detect the expression level of 13 biofilm-associated genes by RT-PCR according to the protocol described previously.25 The fold change of mRNAs expression was calculated using the 2−ΔΔCt method, and the relative expression of each gene was normalized to the control sample (planktonic stage), which was assigned a value of 1 arbitrary unit.26 All assays were performed in triplicate.
Statistical Analysis
Statistical analyses and graphs were performed using GraphPad Prism 8.0.1 (GraphPad Software Inc, USA), Origin 2018 (OriginLab, USA) and SPSS 24.0 (SPSS Statistics, Inc., Chicago, IL, USA). Based on the distribution and homogeneity of the variances, the OD values were expressed as the mean ± SD or median (interquartile range, IQR). Spearman’s rank correlation test was used for intergroup comparison. Mann–Whitney U-test is used to analyze the carrying situation of 13 genes and biofilm formation capacity. Chi-square test was used to compare the positive rates of 13 genes and the number of genes carried in different resistance types of strains. Student’s t-test was used for gene expression in planktonic stage and biofilm stage. In all analyses, a two-sided significance level of <0.05 was considered as statistically significant.
Results
Distribution and Antimicrobial Susceptibility Profiles of Clinical A. baumannii Strains
One hundred and four clinical A. baumannii isolates were collected from 18 departments within the major of intensive care unit (27, 26.2%) and department of neurology (22, 21.4%). Furthermore, the majority of specimens were obtained from sputum, accounting for 72.8%, while only 12.6%, 7.7%, 4.9%, and 2% specimens were isolated from secretion, blood, cerebrospinal fluid, and urine, respectively. The AST results for 18 antimicrobials are shown in Figure 1. Most of isolates (68, 65.4%) were resistance to ciprofloxacin and piperacillin/tazobactam, followed by levofloxacin (67, 64.4%), cefepime (67, 64.4%), meropenem (67, 64.4%), imipenem (67, 64.4%), ceftazidime (67, 64.4%), ceftriaxone (65, 62.5%), cefotaxime (65, 62.5%), piperacillin (62, 59.6%), ampicillin/sulbactam (61, 58.7%), ticarcillin/clavulanic acid (60, 57.7%), gentamicin (54,51.9%), tobramycin (52, 50%), amikacin (52, 50%), and tetracycline (30, 28.8%). More than 65.4% of the isolates exhibited multidrug resistance (Figure 1B), but no strain was resistant to tigecycline and polymyxin B.
Biofilm Formation Ability and Growth Curve Analysis
The biofilm-forming capacity of each isolate was summarized. The OD570 values for the reference strain (ATCC19606) and negative control were 0.84 ± 0.12 and 0.14 ± 0.008, respectively (Table S2). The OD570 values for the clinical isolates ranged from 0.14 ± 0.01 to 2.54 ± 0.26, and the IQR was 0.58 (0.36, 1.04). It is worth noting that 99% of these strains can form biofilm, of which 51.5% were strong biofilm producers. In addition, 35.6% strains produced stronger biofilm than ATCC19606. No significant difference was observed between eight strong and eight weak biofilm producers in growth curves, indicating that the difference in biofilm formation was not due to growth rate (Figure 2). Furthermore, four strains were selected to observe the biofilm formation ability using CLSM. The strongest biofilm-forming strain 20047 reflected the highest fluorescence intensity, followed by strain 20192 and ATCC19606, and biofilm-forming negative strain 19014 presented the lowest fluorescence intensity (Figure 3A). The fluorescence intensities under CLSM were consistent with the OD570 values obtained by crystal violet staining (Figure 3B).
Correlation Between Antimicrobial Resistance and Biofilm Formation Ability
Spearman’s rank correlation analysis determined that non-MDR strains tended to form stronger biofilms than MDR strains (rs= -0.212, P = 0.03; Table 1), suggesting a negative correlation between biofilm formation capacity and antimicrobial resistance phenotypes. Moreover, the results revealed that in addition to tigecycline and polymyxin B, susceptible strains could form stronger biofilm than resistant strains, indicating a negative correlation between biofilm-forming ability and resistance profiles to each of tested antimicrobials (rs= 0.204 − 0.288, P = 0.003 − 0.037; Table 2)
 |
Table 1 Biofilm Formation Ability of Strains with Different Antimicrobial Resistance |
 |
Table 2 Correlation Between Biofilm Formation Ability and Antimicrobial Resistance to 18 Antimicrobials |
Comparison of MICs/MBICs and MBCs/MBECs
The MICs and MBICs of three antimicrobials (ceftazidime, ciprofloxacin, and meropenem) were tested against nine strains with different biofilm-forming ability (three strong biofilm producers, three moderate biofilm producers, and three weak biofilm producers), and all strains were sensitive to these three antibiotics being investigated. MBICs for biofilms increased drastically compared with MIC for planktonic cells among all tested strains (Table 3). The increases ranged from 256- to 4096-fold for ciprofloxacin, 16- to 1024-fold for meropenem, and 128- to 1024-fold for ceftazidime. Indeed, when biofilms were established, the antimicrobial susceptibility profiles of all strains changed from sensitive to resistance.
 |
Table 3 Comparison of MICs and MBICs of A. baumannii Strains |
Furthermore, MBCs and MBECs of these nine isolates were also evaluated. Undoubtedly, the MBECs were much higher than the MBCs among all tested strains (Table 4). Up to a 4096-fold increase in the concentration of ciprofloxacin and up to a 1024-fold increase in the concentration of ceftazidime and meropenem were required to eradicate biofilms compared with planktonic cells.
 |
Table 4 Comparison of MBCs and MBECs of A. baumannii Strains |
Distribution and Expression of Biofilm-Associated Genes in Clinical A. baumannii Strains
In order to further explore the correlation between biofilm-forming ability and antimicrobial resistance, the presence of 13 reported biofilm-associated genes was detected by using PCR. The results showed that bfmR (98, 94.2%) was occurred mostly, followed by csuC (95, 91.3%), csuE (95, 91.3%), bigR (92, 88.5%), csuAB (91, 87.5%), csuB (90, 86.5%), OmpA (89, 85.6%), bfmS (88, 84.6%), bap (85, 81.7%), pgaD (80, 76.9%), csuA (76, 73.1%), csuD (73, 70.2%), and abaI (73, 70.2%). Twenty-five clinical isolates contained all 13 tested genes, including 23 strong biofilm producers and two moderate biofilm producers (Table S3). Remarkably, the biofilm formation ability of the csuD-positive strains was stronger than that of the csuD-negative strains (U = 365, P < 0.01).
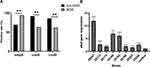
The harboring rates of 13 biofilm-associated genes among two groups were evaluated by Chi-square test. Three genes were significantly related to antimicrobial resistance. In detail, the strains in MDR group had higher carrying rate of csuA and csuD genes than non-MDR group (P < 0.05); however, the non-MDR strains possessed more ompA gene than MDR group (P = 0.002) (Figure 4A).
Eight strong biofilm-forming strains that carrying 13 biofilm-associated genes were selected to investigate the relationship between biofilm-associated genes and biofilm formation ability (Table S4). The RT-PCR results showed that the expression of abaI gene in the biofilm stage was significantly up-regulated compared with that in the planktonic stage (P < 0.001) (Figure 4B), while no significant difference was found for the rest of the 12 genes between planktonic stage and biofilm stage.
Discussion
Drug-resistant A. baumannii shows superior ability to disseminate worldwide, and the proportion of extreme drug resistance (XDR) A. baumannii has increased from less than 4% in 2000 to more than 60% recently, even close to 90% in some regional nosocomial settings.27 According to the National Institutes of Health and the Center for Disease and Prevention, it is estimated that 65% to 80% of the human infectious are caused by biofilm-forming bacteria.28 The lifestyle changes from planktonic phenotype to biofilm is a highly regulated developmental process that depends on species-dependent environmental and genetic factors.4
This study evaluated the biofilm-forming ability of 104 clinical A. baumannii strains, and explored the effect of biofilm production on antimicrobial susceptibility. The MDR strains have reached 65.4%, which increased the difficulty of infection treatment. The AST results revealed that all tested strains remained sensitive to polymyxin B and tigecycline, and it seemed that these two antimicrobials were the most effective strategy for MDR-A. baumannii infections. However, two meta-analyses discourage using tigecycline for the treatment of MDR-A. baumannii infections due to the higher in-hospital mortality and longer hospitalization when compared with other active antimicrobials.29,30 Moreover, polymyxin B rarely used in clinical treatment because of the lack of pharmacokinetic/pharmacodynamic studies and highly renal toxicity.31 Therefore, the optimal treatment for MDR-A. baumannii infections have not been established so far.32
Then, the crystal violet staining results demonstrated that 99% strains have biofilm-forming ability and nearly one-half of strains can produce strong biofilms, which was consistent with the previous reports.24,33 Up to date, there is still no regular pattern in the relationship between biofilm formation ability and bacterial antimicrobial resistance, and the conclusions between different researches were even contradictory.24,33–36 In our research, the non-MDR strains could produce stronger biofilm compared with MDR strains, indicating that the antimicrobial resistance and biofilm formation ability were negatively correlated in A. baumannii. Rodriguez-Bano et al have illustrated that biofilm-forming A. baumannii isolates were more sensitive to ciprofloxacin and imipenem,37 suggesting that these strains were not as dependent on antimicrobial resistance as non-biofilm-forming isolates for survival, which was consistent with our result. Moreover, a research has found that the biofilm-forming ability of ciprofloxacin-sensitive Pseudomonas aeruginosa was significantly promoted after exposure to ciprofloxacin,38 highlighting the importance of biofilms for the survival of sensitive bacteria. Therefore, it seems that the biofilm formation is another survival mechanism besides antimicrobial resistance. Furthermore, MIC/MBIC and MBC/MBEC values confirmed that the resistance of isolates was greatly enhanced after biofilm formation, regardless of whether the isolates were originally sensitive or resistant to this antimicrobial. Remarkably, antimicrobials inhibit biofilms mainly through a complex penetration process,39 however, this still cannot complete remove the bacteria in the biofilm stage due to multifactorial tolerance of biofilms.40 For example, beta-lactam antimicrobials can be inactivated by beta-lactamases in the biofilm matrix, which restricts their penetration. Biofilm-stage bacteria with low metabolic activity can resistant to kinds of antimicrobials that target processes that occur in growing bacteria, such as cell wall synthesis and DNA replication.41 In addition, the expression of specific genes and dormant state persister cells are also responsible for the failure inhibition of biofilms.40,42
In addition to the correlation between antimicrobial resistance and biofilm formation, the relationship between biofilm formation and biofilm-associated genes expression was also evaluated in this study. All the biofilm-associated genes were detected in more than 70% of tested strains, bring with the most frequent gene bfmR (98, 94.2%) as well as the least genes csuD and abaI (70.2%). It has been known that the chaperone usher pili assembly system is essential for the initial steps of biofilm formation.43 Tomaras et al have certified that the inactivation of the csuE gene resulted in the disappearance of both pili formation and biofilm formation in A. baumannii.44 The gene csuD is part of the CsuA/BABCDE chaperone-usher pili assembly system, in this study, csuD-positive strains can form stronger biofilm than csuD-negative strains. This result was consistent with the previous study that indicating csuD was highly expressed in stronger biofilm producers.45 Moreover, in this work, three genes had a obviously distributed regularity among strains with different antimicrobial-resistant profiles. With the increase of resistance, there was a growing positive rate of ompA and a decreasing positive rate of csuA and csuD. Although CsuA/BABCDE system is able to enhance biofilm formation, the relationship between this system and antimicrobial resistance still not clear. OmpA is a major component of outer membrane proteins (OMPs) in A. baumannii, which is related to antimicrobial resistance, aggressiveness, and biofilm formation.13 Smani et al first determined that the deletion of ompA decreased the MICs of chloramphenicol, nalidixic and aztreonam by 8-, 2.67-, and 8-fold, respectively.46 Sanchez-Encinales et al demonstrated that overproduction of OmpA was an independent risk factor for nosocomial pneumonia, bacteremia, and higher mortality caused by A. baumannii infections.47 A previous study confirmed that ompA measured by RT-PCR can be used as a rapid diagnostic factor for antimicrobial-resistant A. baumannii due to the high consistency with MICs.48 In A. baumannii, QS system is consists of AbaI/AbaR system, which is closely interconnected with biofilm development.12 The abaI gene encodes the self-induced synthase which activates the synthesis of AHL signals, and abaI mutant leads a greatly reduction of biofilm formation.12,34 In this study, abaI was the only gene that up-regulated in biofilm stage; however, other tested genes did not show regular changes between planktonic and biofilm stages. A recent study has shown that the absence of AHL signal in shaking cultures (planktonic) seemed to be the result of low expression of abaI, while the AHL signal was significantly up-regulated under static conditions (biofilm).49 Niu et al have demonstrated that the biofilm formation ability of wild type and abaI mutant was no difference at 8 h, but the biofilm formation ability of the mutant was significantly lower than that of wild type at 16 h and 24 h, which was 40% and 41%, respectively. These results suggest that the abaI-directed QS pathway is required for the later stages of biofilm maturation.12 The biofilms were established for 24 h in this study, and it might be the reason why only abaI was overexpressed and the expressions of other genes that contributed to the initial steps of biofilm formation were not significantly changed.1 Indeed, the mechanism of biofilm-associated genes in biofilm formation and antimicrobial resistance needed to be further explored.
Conclusion
In this study, most of A. baumannii have biofilm formation ability and high carrying rate of biofilm-associated genes. There was a negative correlation between biofilm formation ability and antimicrobial resistance, which was stronger biofilm formation occurring in non-MDR strains. Moreover, once the biofilm was formed, whether the biofilm was strong or weak, the antimicrobial resistance can be greatly improved. In addition, presence of csuD was significantly associated with biofilm-forming capacity. The csuA and csuD genes more occured in non-MDR strains, but ompA was more common in MDR strains. Notably, the expression of QS-related gene abaI up-regulated in biofilm stage. In fact, the number and type of biofilm-associated genes carried by biofilm-forming strains are quite different among different regions,11,50,51 indicating that the biofilm-forming ability is not only determined by the presence of biofilm-associated genes but a complex process depended on multiple factors. Deeper exploration of mechanisms of biofilm formation and biofilm-specific resistance in A. baumannii are highly needed.
Ethics Approval and Consent to Participate
This study was approved by the Institutional Review Board of affiliated hospital of southwest medical university (KY2020043). Written informed consent was obtained from all participants.
Acknowledgments
This work was supported by the grants from Sichuan Science and Technology Program (2021YFH001, 21ZDYF2148, 2019YFS0038), Southwest Medical University Science Program (2020ZRQNB025), and Luxian Government and Southwest Medical University Cooperation Program (2020LXXNYKD-04).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. doi:10.1038/nrmicro.2017.148
2. Greene C, Wu J, Rickard AH, Xi C. Evaluation of the ability of Acinetobacter baumannii to form biofilms on six different biomedical relevant surfaces. Lett Appl Microbiol. 2016;63(4):233–239. doi:10.1111/lam.12627
3. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi:10.1038/nrmicro2415
4. Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3):276–301.
5. Amin M, Navidifar T, Shooshtari FS, et al. Association between biofilm formation, structure, and the expression levels of genes related to biofilm formation and biofilm-specific resistance of Acinetobacter baumannii strains isolated from burn infection in Ahvaz, Iran. Infect Drug Resist. 2019;12:3867–3881. doi:10.2147/IDR.S228981
6. Brossard KA, Campagnari AA. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun. 2012;80(1):228–233. doi:10.1128/IAI.05913-11
7. Luo LM, Wu LJ, Xiao YL, et al. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol. 2015;15:62. doi:10.1186/s12866-015-0397-5
8. Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology (Reading, England). 2008;154(Pt 11):3398–3409. doi:10.1099/mic.0.2008/019471-0
9. Pakharukova N, Tuittila M, Paavilainen S, et al. Structural basis for Acinetobacter baumannii biofilm formation. Proc Natl Acad Sci USA. 2018;115(21):5558–5563. doi:10.1073/pnas.1800961115
10. Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litrán T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol. 2009;191(19):5953–5963. doi:10.1128/JB.00647-09
11. Shenkutie AM, Yao MZ, Siu GK, Wong BKC, Leung PH. Biofilm-induced antibiotic resistance in clinical Acinetobacter baumannii isolates. Antibiotics (Basel, Switzerland). 2020;9(11). doi:10.3390/antibiotics9110817
12. Niu C, Clemmer KM, Bonomo RA, Rather PN. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol. 2008;190(9):3386–3392. doi:10.1128/JB.01929-07
13. Nie D, Hu Y, Chen Z, et al. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J Biomed Sci. 2020;27(1):26. doi:10.1186/s12929-020-0617-7
14. Walsh BJC, Wang J, Edmonds KA, et al. The response of Acinetobacter baumannii to hydrogen sulfide reveals two independent persulfide-sensing systems and a connection to biofilm regulation. mBio. 2020;11(3). doi:10.1128/mBio.01254-20.
15. Clinical and Laboratory Standards Institute; Wayne PA. Performance Standards for Antimicrobial Susceptibility Testing, 30th Edition. CLSI M100. CLSI; 2020.
16. Ramirez MS, Bonomo RA, Tolmasky ME. Carbapenemases: transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules. 2020;10(5):720. doi:10.3390/biom10050720
17. Krishnamoorthy S, Shah BP, Lee HH, Martinez LR. Microbicides alter the expression and function of RND-type efflux pump AdeABC in biofilm-associated cells of Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. 2016;60(1):57–63. doi:10.1128/AAC.01045-15
18. Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–179. doi:10.1016/S0167-7012(00)00122-6
19. Pakkulnan R, Anutrakunchai C, Kanthawong S, Taweechaisupapong S, Chareonsudjai P, Chareonsudjai S. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One. 2019;14(3):e0213288. doi:10.1371/journal.pone.0213288
20. Zhang B, Yu P, Wang Z, Alvarez PJJ. Hormetic promotion of biofilm growth by polyvalent bacteriophages at low concentrations. Environ Sci Technol. 2020;54(19):12358–12365. doi:10.1021/acs.est.0c03558
21. Tang M, Wei X, Wan X, Ding Z, Ding Y, Liu J. The role and relationship with efflux pump of biofilm formation in Klebsiella pneumoniae. Microb Pathog. 2020;147:104244. doi:10.1016/j.micpath.2020.104244
22. Moskowitz SM, Foster JM, Emerson J, Burns JL. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 2004;42(5):1915–1922. doi:10.1128/JCM.42.5.1915-1922.2004
23. Gao B, Li X, Yang F, et al. Molecular epidemiology and risk factors of ventilator-associated pneumonia infection caused by carbapenem-resistant enterobacteriaceae. Front Pharmacol. 2019;10:262. doi:10.3389/fphar.2019.00262
24. Yang CH, Su PW, Moi SH, Chuang LY. Biofilm formation in Acinetobacter baumannii: genotype-phenotype correlation. Molecules (Basel, Switzerland). 2019;24(10). doi:10.3390/molecules24101849
25. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi:10.1373/clinchem.2008.112797
26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
27. Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30(1):409–447. doi:10.1128/CMR.00058-16
28. Wolcott R, Dowd S. The role of biofilms: are we hitting the right target? Plast Reconstr Surg. 2011;127(Suppl 1):28s–35s. doi:10.1097/PRS.0b013e3181fca244
29. Ni W, Han Y, Zhao J, et al. Tigecycline treatment experience against multidrug-resistant Acinetobacter baumannii infections: a systematic review and meta-analysis. Int J Antimicrob Agents. 2016;47(2):107–116. doi:10.1016/j.ijantimicag.2015.11.011
30. Kengkla K, Kongpakwattana K, Saokaew S, Apisarnthanarak A, Chaiyakunapruk N. Comparative efficacy and safety of treatment options for MDR and XDR Acinetobacter baumannii infections: a systematic review and network meta-analysis. J Antimicrob Chemother. 2018;73(1):22–32. doi:10.1093/jac/dkx368
31. Cai Y, Lee W, Kwa AL. Polymyxin B versus colistin: an update. Expert Rev Anti Infect Ther. 2015;13(12):1481–1497. doi:10.1586/14787210.2015.1093933
32. Garnacho-Montero J, Timsit JF. Managing Acinetobacter baumannii infections. Curr Opin Infect Dis. 2019;32(1):69–76. doi:10.1097/QCO.0000000000000518
33. Qi L, Li H, Zhang C, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7:483. doi:10.3389/fmicb.2016.00483
34. Gaddy JA, Actis LA. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009;4(3):273–278. doi:10.2217/fmb.09.5
35. Khoshnood S, Savari M, Abbasi Montazeri E, Farajzadeh Sheikh A. Survey on genetic diversity, biofilm formation, and detection of colistin resistance genes in clinical isolates of Acinetobacter baumannii. Infect Drug Resist. 2020;13:1547–1558. doi:10.2147/IDR.S253440
36. Lin MF, Lin YY, Lan CY. Characterization of biofilm production in different strains of Acinetobacter baumannii and the effects of chemical compounds on biofilm formation. PeerJ. 2020;8:e9020. doi:10.7717/peerj.9020
37. Rodríguez-Baño J, Martí S, Soto S, et al. Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin Microbiol Infect. 2008;14(3):276–278. doi:10.1111/j.1469-0691.2007.01916.x
38. Shi N, Gao Y, Yin D, et al. The effect of the sub-minimal inhibitory concentration and the concentrations within resistant mutation window of ciprofloxacin on MIC, swimming motility and biofilm formation of Pseudomonas aeruginosa. Microb Pathog. 2019;137:103765. doi:10.1016/j.micpath.2019.103765
39. Stewart PS. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40(11):2517–2522. doi:10.1128/AAC.40.11.2517
40. Ciofu O, Rojo-Molinero E, Macià MD, Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125(4):304–319. doi:10.1111/apm.12673
41. Jensen P, Briales A, Brochmann RP, et al. Formation of hydroxyl radicals contributes to the bactericidal activity of ciprofloxacin against Pseudomonas aeruginosa biofilms. Pathog Dis. 2014;70(3):440–443. doi:10.1111/2049-632X.12120
42. De Groote VN, Verstraeten N, Fauvart M, et al. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol Lett. 2009;297(1):73–79. doi:10.1111/j.1574-6968.2009.01657.x
43. McQueary CN, Actis LA. Acinetobacter baumannii biofilms: variations among strains and correlations with other cell properties. J Microbiol (Seoul, Korea). 2011;49(2):243–250. doi:10.1007/s12275-011-0343-7
44. Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology (Reading, England). 2003;149(12):3473–3484. doi:10.1099/mic.0.26541-0
45. Selasi GN, Nicholas A, Jeon H, et al. Differences in biofilm mass, expression of biofilm-associated genes, and resistance to desiccation between epidemic and sporadic clones of carbapenem-resistant Acinetobacter baumannii sequence type 191. PLoS One. 2016;11(9):e0162576. doi:10.1371/journal.pone.0162576
46. Smani Y, Fàbrega A, Roca I, Sánchez-Encinales V, Vila J, Pachón J. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58(3):1806–1808. doi:10.1128/AAC.02101-13
47. Sánchez-Encinales V, Álvarez-marín R, Pachón-Ibáñez ME, et al. Overproduction of outer membrane protein A by Acinetobacter baumannii as a risk factor for nosocomial pneumonia, bacteremia, and mortality rate increase. J Infect Dis. 2017;215(6):966–974. doi:10.1093/infdis/jix010
48. Martín-Peña R, Domínguez-Herrera J, Pachón J, McConnell MJ. Rapid detection of antibiotic resistance in Acinetobacter baumannii using quantitative real-time PCR. J Antimicrob Chemother. 2013;68(7):1572–1575. doi:10.1093/jac/dkt057
49. Mayer C, Muras A, Romero M, López M, Tomás M, Otero A. Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front Cell Infect Microbiol. 2018;8:310. doi:10.3389/fcimb.2018.00310
50. Altınok Ö, Boral B, Ergin A, Eser ÖK. Existence of biofilm and biofilm-associated virulence genes in multi-drug resistant invasive Acinetobacter baumannii isolates. Mikrobiyol Bul. 2020;54(1):40–49. doi:10.5578/mb.20204
51. Liu H, Wu YQ, Chen LP, et al. Biofilm-related genes: analyses in multi-antibiotic resistant Acinetobacter baumannii isolates from Mainland China. Medical Sci Monit. 2016;22:1801–1807. doi:10.12659/MSM.898959
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.