Back to Journals » Journal of Inflammation Research » Volume 14
Phenotypes, Lung Microbiota and Cytokine Responses in Pneumonia After Hematopoietic Stem Cell Transplantation
Authors Hu Y, Jiang Y, Liu S, Shen J , An Y
Received 12 September 2021
Accepted for publication 12 November 2021
Published 19 November 2021 Volume 2021:14 Pages 6055—6065
DOI https://doi.org/10.2147/JIR.S338914
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Yan Hu,1 Yanwen Jiang,1 Shuang Liu,1 Jiawei Shen,2 Youzhong An2
1Department of Respiratory and Critical Care Medicine, Peking University International Hospital, Beijing, People’s Republic of China; 2Department of Critical Care Medicine, Peking University People’s Hospital, Beijing, People’s Republic of China
Correspondence: Jiawei Shen; Youzhong An
Department of Critical Care Medicine, Peking University People’s Hospital, Beijing, People’s Republic of China
Email [email protected]; [email protected]
Objective: We aim to identify phenotypes of hematopoietic stem cell transplantation (HSCT) patients with pneumonia, discover relations of microbiota composition, cytokine profile, and outcomes between phenotypes. Specific cytokines will be evaluated for their role in lung injury in a murine model.
Methods: HSCT patients with pneumonia were included, and clustering of variables including cytokine levels provided the phenotypes. Outcomes were compared between phenotypes. Analysis of lung microbiota identified marker species of phenotypes. In the murine model, marker species-related cytokine regulations and the role of cytokines in lung injury were evaluated.
Results: Seventy-two patients were included, and two phenotypes were identified, namely “reactive” (N=21) and “nonreactive” (N=51) phenotype. Compared to their counterparts, patients with nonreactive phenotype had lower serum IL-6, IL-8, less severe inflammation, worse outcomes and more viruses as marker species in lung microbiota. The animal study validated the pathogens specific cytokine responses that presented in the human study and the potential protective role of IL-6 in these patients.
Conclusion: HSCT patients with pneumonia can be clustered into two phenotypes with different marker species and outcomes: the “nonreactive” phenotype and the “reactive” phenotype. Serum cytokine levels were different between the two phenotypes, which indicate the existence of the pathogen-related cytokine responses. For patients with the “nonreactive” phenotype, IL-6 therapy may improve their prognosis, which should be further tested in clinical studies.
Keywords: hematopoietic stem cell transplantation, pneumonia, phenotype, microbiota, cytokines
Introduction
Hematopoietic stem cell transplantation (HSCT) is widely applied and validated as a cure for many hematological diseases and many other disorders.1 Although the prognosis of HSCT has improved with the advance of the treatment strategies,2 various complications including pneumonia still had a high prevalence and mortality, Lucena et al reported that LTRI had an incidence of 68%,3 1-year mortality of these patients were 24–52.6% in different studies.3,4
Substantial heterogeneity within HSCT patients may have contributed to the various prognoses and responses to treatments of pneumonia. For example, in HSCT patients with influenza pneumonia, the reported mortality ranged from 6.3% to 25%,5,6 mortality of Respiratory Syncytial virus pneumonia was 29% to 88%.7–9 For those who need respiratory supports, their response to mechanical ventilation10 and extracorporeal membrane oxygenation11 also differs between patients with different characteristics. Recently, Calfee et al and Bos et al have identified phenotypes of acute respiratory distress syndrome (ARDS) based on clinical and biological data (including serum cytokine levels), patients of different phenotypes have distinct presentations of cytokine levels, other inflammation presentations, clinical outcomes and response to specific treatments.12–14
Progress in next-generation sequencing and related bioinformatics analysis has provided a new perspective on this type of patient. Based on sequencing data from bronchial alveolar lavage fluid (BALF) samples, Herivaux et al discovered that a decreased alpha diversity increased abundance of a specific genus of bacteria was related to a higher risk of invasive pulmonary aspergillosis in HSCT patients.15 O’Dwyer et al reported that in HSCT patients, alveolar cytokine levels were associated with the features of lung microbiomes, besides, patients with lower diversity of lung microbiota had higher incidence of pulmonary complications.16
We then hypothesized that 1) there are phenotypes within HSCT patients with pneumonia, outcomes of patients will differ between different phenotypes. 2) the composition of microbiota and cytokine levels differ between phenotypes 3) Specific cytokines that expressed differently within each phenotype would affect the severity of lung injury induced by infection, which could be potential therapeutic targets in future studies.
Methods
Ethics Statement
The clinical study was approved by the Ethics and Research board of Peking University People’s Hospital (PKUPH, 2020PHB400-01). The clinical study was conducted under the principles of the Helsinki Declaration, informed consents were collected from patients or their relatives before participation. The animal study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals by the NIH and approved by the Animal Center of Peking University Health Science Center.
Human Study
In the human study, HSCT patients with pneumonia were divided into different phenotypes with hierarchical clustering, outcomes of patients with different phenotypes were compared. Lung microbiotas were analyzed to find marker families and species for different phenotypes.
Patients in Human Study
We included patients who were admitted to the Intensive Care Units of PKUPH and PKUIH (from August 2020 to July 2021) with a diagnosis of HSCT and pneumonia. Definition of pneumonia was as follows: 1) Have at least one respiratory symptom (sputum production, cough, dyspnea, tachypnea, pleuritic pain) and 2) Abnormal auscultation (rales or crepitation) or at least one sign of infection (Temperature >38.0°C, shivering, white blood cell count >10 ×109/L or <4000 ×109/L) independent of antibiotic therapy and 3) New infiltrate on chest radiographs.17,18 Exclusion criteria: We excluded patients 1) <18 years old, 2) in pregnancy and 3) with a diagnosis of diffuse alveolar hemorrhage, asthma, bronchiectasis or chronic obstructive pulmonary disease. Baseline characteristics (eg, age, sex and days after HSCT) and clinical outcomes (Ventilator free days and 30-day mortality) of patients were recorded.
Blood Sampling, Laboratory Tests and Cytokine Analysis
On the day of ICU admission, blood samples were collected from included patients, regular laboratory tests including whole blood count, blood chemistry tests (ie, creatine, lactate), C-reactive protein, procalcitonin levels, coagulation functions (ie prothrombin time) and arterial blood gas (ie, PO2/FiO2) were performed in the hospital laboratory. Cytokine levels (IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IFN-γ, TNF-α) were measured with a magnetic bead array panel (Cytokine Magnetic 10-Plex Panel) in accordance with the instruction of the manufacturer (Thermo Fisher, Waltham, MA, USA).
Hierarchical Clustering of Phenotypes, Characteristics and Outcomes of Different Phenotypes
Clinical characteristics and cytokine data were normalized, and Ward’s hierarchical clustering was performed to minimize the variance within each cluster (with “ggdendro” package in R). The optimal number of clusters was determined using “Nbclust” package. The clustering result was visualized with a dendrogram and heatmap using “factoextra” and “ggplot2” packages. Characteristics and outcomes were compared between different phenotypes. Kaplan–Meier statistics were used to estimate the 30-day survival of patients striated by phenotypes was conducted and visualized with “survival” and “survminer” packages.
Bronchial Alveolar Lavage Sampling and RNA-Based Metagenomic Next-Generation Sequencing
All included patients received a bronchoscope examination with bronchial alveolar lavage within the first 48 hours of ICU admission. At least 5 mL of BALF was collected for each patient. BALF samples were analyzed with an RNA-based metagenomic next-generation sequencing (mNGS) to ensure the detection of RNA viruses along with DNA viruses, bacteria and fungus. The process was performed as previously described.19 Briefly, RNA was extracted from BALF samples. Through reverse transcription, cDNA libraries were constructed and sequenced with NextSeq 550 System (150-bp paired-end reads; Illumina) to produce sequencing data for further analysis.
Bioinformatic Analysis of Sequencing Data
Sequencing data were processed with “Kneaddata” (v0.9.0) to remove host contaminations.20 Cleaned data were analyzed with “Kraken2” (v2.1.2) to assign taxonomy in reference to a database based on NCBI library.21 We used “Bracken” (Bayesian Reestimation of Abundance with KrakEN, v2.1.0) to calculate the abundances of all species detected in the last step.22 To identify species or families as markers associated with every phenotype, linear discriminant analysis effect size (LDA with LEfSe, v1.1.1) was applied to the reformatted Bracken output data. The visualization of alpha diversity, beta diversity and stack-plot of species contribution were achieved using the “EazyAmplicon” pipeline23 in R.
Etiological Diagnosis for Patients
The clinical etiological diagnoses of the included patients were made based on next-generation sequencing tests, pathogen culture results of bronchoalveolar lavage fluids, and with reference to radiological features of the patient. Then, the doctor in charge of the patient will classify pneumonia as viral, bacterial, fungal or with other causes, and the suspected pathogens for all patients were also recorded.
Animal Study
Based on the result of the human study, the animal study was designed to further verify the existence of marker species (pathogens)-related cytokine regulations. Additionally, to clarify the effect of the related cytokines on lung injury.
Syngeneic Bone Marrow Transplantation for Mice
IL-6 knockout (KO) and wild-type (WT) C57 BL/6 mice (Jackson Laboratory, Bar Harbor, ME, USA) were used in the animal study. Syngeneic bone marrow transplantation (BMT) was performed as previously described,24 except that we only perform transplantation between IL-6 KO (donor), WT (donor) and WT (recipients); thus, there are two types of mice after BMT: IL-6 → WT and WT→ WT. Mice will not be infected until the fifth week after transplantation when the hematopoietic cells are reconstructed.25
Intervention Protocol
Pseudomonas Aeruginosa (PA) and Murine gammaherpesvirus 68 (MHV) were used as agents of infection in mice after BMT, IL-6 treatment was applied to clarify the role of IL-6 to lung injury. PA and MHV infections were performed as previously described.26,27 For the IL-6 treatment, mice were treated intranasally with mouse recombinant IL-6 protein (R&D, Minneapolis, MN) at a dosage of 25ug per mouse, diluted with PBS with 0.1% BSA to 50 µL, intranasal inoculated on day 4, 5, 6, 7 post infection.28 BALF samples and lung tissues were harvested at day 8 post-infection.
BALF Protein and Histological Analysis
We analyzed protein and IL-6 concentration of BALF using the Modified Lowry Method Protein Assay Kit (Sigma Aldrich, St. Louis, MO, USA) and a Mouse IL-6 Quantikine ELISA Kit (R&D, Minneapolis, MN), according to the instructions of the manufacturer, respectively.
For histological examination, the whole lung was perfused with 500 ul of 4% neutral paraformaldehyde in situ and fixed for 24 hours in 4% neutral paraformaldehyde. Five-micrometer sections from a fixed, paraffin-embedded whole lung were then stained with hematoxylin and eosin. Immunochemical labelling of IL-6 was performed as previously described.29
Statistical Analysis
Estimation of sample size was achieved using PASS (V11.0.7, NCSS, Kaysville, UT, USA). The reported 30-day mortality for HSCT patients with respiratory failure was 50–75%.10,30 To present the estimated mortality rate in the study cohort (62.5% as a mean value), we set the allowable error rate as 6.25% (62.5% × 0.1), confidence interval as 0.9, a sample size of 65 is estimated. All statistical analyses were performed on R (v4.0.4). The missing variables were replaced using multiple imputation chained equations (“MICE” package). Variables were presented as mean ± standard deviation or median (interquartile range) as appropriate, categorical variables were presented as number (percentage). Comparison of two groups of continuous variables was performed with the Student’s t-test or Mann–Whitney U-test as appropriate. Comparison of a categorical variable (ie 30-day mortality) was achieved using χ2 test. Correlations between categorical variables were evaluated with Phi coefficient or Cramer’s V coefficient as appropriate. All tests were two-sided and p < 0.05 was considered statistically significant.
Results
Patients
One hundred and five patients with a diagnosis of HSCT were admitted into the two centers during the study period. Eighty-one patients were also diagnosed with pneumonia; among them, 9 patients meet the exclusion criteria, and the rest 72 patients were included in the study. The baseline characteristics of patients are reported in Table 1. Most of the patients received allogeneic HSCT (70[97.2%]), 34 (47.22%) patients received stem cells from donors with haploidentical HLA matching.
 |
Table 1 Baseline Characteristics of Patients |
Identification of Phenotypes
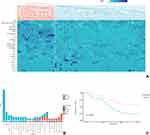
Two clusters were separated based on clinical characteristics, laboratory tests and serum cytokine levels (Figure 1A). We defined the first cluster as the “reactive” phenotype (21 patients), and the second cluster as the “nonreactive” phenotype (51 patients). When compared to patients with nonreactive phenotype, patients with reactive phenotype had higher PaO2/FiO2 (Table 2) in arterial blood gas, higher max temperature, white blood cells count and granulocyte count, higher C-reactive protein (CRP) and creatine levels, higher serum IL-6 and IL-8 levels.
 |
Table 2 Characteristics and Outcomes of Patients with Different Phenotypes |
Etiological Diagnosis for Patients
Details of suspected pathogens for patients were recorded in Table S1 of Supplementary Files, where many pathogens mutually existed in the two phenotypes. Viral pathogens contributed to a large proportion of the nonreactive phenotypes, while bacterial pathogens were more often identified in patients with the reactive phenotype (Figure 1B). CMV was moderately correlated with the nonreactive phenotype (Phi=0.370, P=0.02) and Pseudomonas Aeruginosa was moderately correlated with the reactive phenotype (Phi=0.379, P=0.001). The overall correlation between pathogens and phenotypes were strong (Cramer’s V=0.504, P<0.001) (Table S2, Supplementary Files).
Worse Outcomes of Patients with Nonreactive Phenotype
Patients with nonreactive phenotype had a higher rate of 30-day mortality (78.43% vs 42.87%, p=0.0035) and lower ventilator-free days (4.25±2.21 vs 6.52±5.65, p=0.0161). Figure 1C depicts the survival curve of patients with reactive or nonreactive phenotypes. Patients with nonreactive phenotype had a significantly lower survival rate (Log-rank p=0.0028).
Microbiota Characteristics and Marker Species of Phenotypes
The alpha diversity, or the diversity within each sample, of patients within reactive or nonreactive phenotype, was evaluated with Shannon’s Diversity Index. Patients with reactive phenotype had higher richness and evenness of species between samples. The beta diversity was evaluated using the Bray-Curtis Dissimilarity metric, which considers the identity and abundance of species shared between phenotypes. This metric was plotted in a principal coordinate analysis (PCoA) and revealed an apparent pattern of clustering (see Figure S1 of Supplementary Material).
The contribution of strains in the samples of the 2 phenotypes is depicted in Figure 2A. In the reactive phenotype, the most abundant species of is Pseudomonas Aeruginosa, while in the nonreactive phenotype, Human Cytomegalovirus has the highest abundance. The species or family profile of the two phenotypes were analyzed by LEfSe (Figure 2B and C), which revealed an enrichment of various bacteria (eg, Pseudomonas Aeruginosa, Klebsiella Pneumoniae, Stenotrophomonas sp. ASS1) as the marker (LDA score >2) species in the reactive phenotype. In the nonreactive phenotype, the maker species were mainly DNA or RNA viruses (eg, Human Cytomegalovirus, Human Respiratory Syncytial Virus, Influenza C virus) and Fungus (eg, Aspergillus Fumigatus) or bacteria (eg, Prevotella Dentalis, Schaalia Odontolytica). On the family levels, markers for reactive phenotype are Pseudomonadaceae, Enterobacteriaceae, Staphylococcaceae and Moraxellaceae. For nonreactive phenotypes, the families are Herpesviridae, Orthomyxoviridae and Pneumoviridae.
MHV Infection is Related with Lower Concentration of BALF IL-6 and More Severe Lung Injury
Based on the results of the human study, different pathogens (marker species) were related to two phenotypes with different IL-6 and IL-8 levels. In the animal study, MHV, a strain in the same subfamily of betaherpesvirinae as the cytomegalovirus, and PA were used as the pathogens. Because mice do not have the IL-8 gene, in the mice model we focused on the pathogen specific IL-6 response and its effect on lung injury.
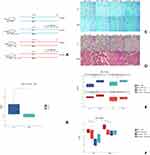
Figure 3A illustrates the details of interventions. For the WT→ WT BMT mice infected with PA, the IL-6 level in BALF was significantly higher than those infected with MHV (Figure 3B), while the immunohistochemical staining also showed a higher number of IL-6 positive cells in PA-infected mice than MHV infected ones (Figure 3C). In the histological study (Figure 3D), lung injuries caused by MHV infection were more serious than in PA infections. There was a consistent trend of higher concentration of BALF protein (Figure 3E) in MHV infected mice than PA infected ones (except for the IL-6 KO → WT + IL-6 group).
IL-6 Treatments Alleviate Lung Injury in BMT Mice That Received IL-6 KO Marrow
In Figure 3C, the difference of IL-6 expression as influenced by BMT and infection was depicted. Mice that received IL-6 KO bone marrows (IL-6 KO → WT) had no IL-6 proteins in cells of lung tissue, while IL-6 distribution was observed in mice treated with IL-6 protein (IL-6 KO → WT + IL-6).
As shown in Figure 3D and F, IL-6 KO → WT mice had much more severe lung injury (caused by PA or MHV) than WT → WT mice, and IL-6 treatment alleviated the severity of the injury. For WT → WT mice, the protective effect of IL-6 did not exist.
Discussion
In this study, we identified two phenotypes in HSCT patients with pneumonia, namely: reactive and nonreactive phenotypes. Compared to their “reactive” counterparts, patients with nonreactive phenotype were characterized with less obvious presentations of inflammation, lower levels of serum IL-6, IL-8 and worse clinical outcomes, their lung microbiota were characterized with lower biodiversity and marked with more virus species. In the murine model, we further validated this marker species (pathogens)-related cytokine responses (ie IL-6). Additionally, the potential protective role of IL-6 in infection-induced lung injury was observed, which could be a potential therapeutic target in HSCT patients with pneumonia that presented as a nonreactive phenotype.
To our knowledge, this is the first study that focused on the phenotypes of HSCT patients with pneumonia. With latent class analysis of data from clinical trials, Calfee et al have identified two phenotypes of ARDS in the general population,12 the “hyperinflammatory” phenotype is characterized by inflammation (including a higher concentration of serum IL-6, IL-8), increased mortality and decreased ventilator-free days. Similar result is reported by Bos et al, and in their study, patients characterized by greater inflammation (higher serum cytokine levels) had increased mortality.13 However, in the present study, a reactive phenotype is related to lower hospital mortality and ventilator-free days. One possible explanation is that ARDS is a clinical syndrome with various etiologies: in addition to lung infections, pancreatitis and sepsis are also common causes associated with hyperinflammatory states.31,32 Yet, patients in our cohort were immunocompromised, and the inflammatory response could not be as competent as the normal subjects. Patients with reactive or nonreactive phenotypes had relatively low SOFA scores (Table 2). And unlike the immunocompetent patients with sepsis, cytokine (eg IL-6) levels in our cohort were much lower (with 95% confidence interval ranging from 76.23 to 550.01 pg/mL for the reactive phenotype vs 453.85–9163.64 pg/mL33 or 243–1295134 pg/mL in sepsis patients). This evidence indicates that the reactive phenotype may not definitely be related to a catastrophic multiple organ failure, as the “reactive phenotype” in our cohort were relatively static in cytokine response when compared to sepsis patients. However, for patients in the “nonreactive phenotype”, their inflammatory response might be insufficient and contribute to worse outcomes. The underlying mechanism for pathogen-related cytokine responses in HSCT patients needs to be clarified in further studies.
Lower alpha diversity was observed in nonreactive patients, and as the previous study reported, lower-alpha diversity of lung microbiota was related to higher mortality of patients after HSCT,16 which was confirmed in our study. Our study also identified the marker species of the two pneumonia phenotypes in HSCT patients and further evaluated this relation in the murine model. The effect of etiologic pathogens and inflammatory responses that were evaluated with cytokine levels were explored in several studies. Burgmeijer et al reported that in patients with community-acquired pneumonia, serum IL-6, IL-17a, and INF-γ were significantly higher in pneumococcal infected ones than those with a strict viral etiology.35 Nonetheless, a higher level of cytokines in bacterial pneumonia (compared to viral pneumonia) was not observed in another study.36 Döring et al reported that compared to viremia, pediatric HSCT patients with bacteremia had significantly higher levels of serum IL-8, but IL-6 levels were similar between the two groups.37 In addition, pneumonia with a specific virus origin (eg Influenza A or SARS-CoV-2) can also cause upregulated cytokine profiles.38,39 With limited evidence, we can only infer whether those specific pathogens (eg, Cytomegalovirus, Pseudomonas Aeruginosa) can influence cytokine regulation in HSCT patients.
Many studies have evaluated the role of IL-6 in infection-related lung injury. In studies of experimental ARDS induced by aspiration of bacterial endotoxin, the addition of recombinant IL-6 reduced pulmonary inflammation40,41 as we discovered. Similarly, IL-6 KO mice that were infected with influenza had higher mortality and reduced macrophage phagocytic function.42,43 This protective role of IL-6 was also observed in mice with HSV-1 pneumonia.44 However, in the model of extrapulmonary, sepsis-induced ARDS, inhibition of IL-6 receptor with tocilizumab reduced lung inflammation and mortality.45 In critically ill patients with coronavirus disease 2019 (COVID-19), IL-6 receptor antagonists Tocilizumab and Sarilumab improved patient survival.46 In the present study, IL-6 treatment alleviated lung injury in recipients of IL-6 KO marrows; nonetheless, recipients of WT marrows did not further benefit from it. With reference to previous studies, the protective or exacerbating role of IL-6 may be contextual, depending on the pathogen’s ability to ignite the inflammatory response and the extent of immune response from the host, in other words, whether the immunological responses are “adaptive” to the infection.47 One analogous situation is the inflammation response in sepsis, where the early stage of sepsis is characterized by a hyper-inflammation stage that is followed by an immunosuppressive stage. Both of the two “maladaptive” immunological statuses contributed to the mortality of sepsis, while patients with a relatively adaptive response had a higher chance of survival.48 Our study indicated that HSCT patients with pneumonia are presenting with two different inflammation phenotypes (reactive, nonreactive). For patients with the “nonreactive” phenotype (possibly with a maladaptive response), IL-6 therapy may improve their prognosis, which should be further tested in clinical studies.
Our study has several limitations. First, although two phenotypes were successfully identified, the sample size of the human study was still limited; this would potentially reduce the reliability of the results. This problem was indirectly solved with validation in the animal study. Second, the animal study did not clarify the mechanisms of pathogens related cytokine (ie, IL-6) responses, also, not all marker species were evaluated in animal studies. Besides, as the application of IL-6 or its monoclonal antibody was off-label in our cohort, their effect in the clinical environment cannot be evaluated in this study. All these issues should be targeted in future studies.
Conclusion
HSCT patients with pneumonia can be clustered into two phenotypes with different marker species and outcomes: the “nonreactive” phenotype and the “reactive” phenotype. Serum cytokine levels were different between the two phenotypes, indicating the existence of the pathogen-related cytokine responses. For patients with the “nonreactive” phenotype, IL-6 therapy may improve their prognosis, which should be further tested in clinical studies.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure
The authors report no conflict of interests.
References
1. Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813–1826. doi:10.1056/NEJMra052638
2. Gooley TA, Chien JW, Pergam SA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363(22):2091–2101. doi:10.1056/NEJMoa1004383
3. Lucena CM, Torres A, Rovira M, et al. Pulmonary complications in hematopoietic SCT: a prospective study. Bone Marrow Transplant. 2014;49(10):1293–1299. doi:10.1038/bmt.2014.151
4. Aguilar-Guisado M, Jimenez-Jambrina M, Espigado I, et al. Pneumonia in allogeneic stem cell transplantation recipients: a multicenter prospective study. Clin Transplant. 2011;25(6):E629–38. doi:10.1111/j.1399-0012.2011.01495.x
5. Ljungman P, Ward KN, Crooks BN, et al. Respiratory virus infections after stem cell transplantation: a prospective study from the Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation. Bone Marrow Transplant. 2001;28(5):479–484. doi:10.1038/sj.bmt.1703139
6. Ljungman P, de la Camara R, Perez-Bercoff L, et al. Outcome of pandemic H1N1 infections in hematopoietic stem cell transplant recipients. Haematologica. 2011;96(8):1231–1235. doi:10.3324/haematol.2011.041913
7. Ljungman P. Respiratory virus infections in bone marrow transplant recipients: the European perspective. Am J Med. 1997;102(3A):44–47. doi:10.1016/s0002-9343(97)00010-7
8. Harrington RD, Hooton TM, Hackman RC, et al. An outbreak of respiratory syncytial virus in a bone marrow transplant center. J Infect Dis. 1992;165(6):987–993. doi:10.1093/infdis/165.6.987
9. Ebbert JO, Limper AH. Respiratory syncytial virus pneumonitis in immunocompromised adults: clinical features and outcome. Respiration. 2005;72(3):263–269. doi:10.1159/000085367
10. Shen J, Hu Y, Zhao H, et al. Risk factors of non-invasive ventilation failure in hematopoietic stem-cell transplantation patients with acute respiratory distress syndrome. Ther Adv Respir Dis. 2020;14:1753466620914220. doi:10.1177/1753466620914220
11. Wohlfarth P, Beutel G, Lebiedz P, et al. Characteristics and outcome of patients after allogeneic hematopoietic stem cell transplantation treated with extracorporeal membrane oxygenation for acute respiratory distress syndrome. Crit Care Med. 2017;45(5):e500–e507. doi:10.1097/CCM.0000000000002293
12. Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2(8):611–620. doi:10.1016/S2213-2600(14)70097-9
13. Bos LD, Schouten LR, van Vught LA, et al. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax. 2017;72(10):876–883. doi:10.1136/thoraxjnl-2016-209719
14. Calfee CS, Delucchi KL, Sinha P, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med. 2018;6(9):691–698. doi:10.1016/S2213-2600(18)30177-2
15. Herivaux A, Willis JR, Mercier T, et al. Lung microbiota predict invasive pulmonary aspergillosis and its outcome in immunocompromised patients. Thorax. 2021:
16. O’Dwyer DN, Zhou X, Wilke CA, et al. Lung dysbiosis, inflammation, and injury in hematopoietic cell transplantation. Am J Respir Crit Care Med. 2018;198(10):1312–1321. doi:10.1164/rccm.201712-2456OC
17. Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi:10.1086/511159
18. Woodhead M. New guidelines for the management of adult lower respiratory tract infections. Eur Respir J. 2011;38(6):1250–1251. doi:10.1183/09031936.00105211
19. Chen H, Yin Y, Gao H, et al. Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin Infect Dis. 2020;71(Suppl 4):S416–S426. doi:10.1093/cid/ciaa1516
20. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962–968. doi:10.1038/s41592-018-0176-y
21. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi:10.1186/gb-2014-15-3-r46
22. Lu JBF, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;2017(1):e104. doi:10.7717/peerj-cs.104
23. Liu YX, Qin Y, Chen T, et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 2021;12(5):315–330. doi:10.1007/s13238-020-00724-8
24. Hamzic N, Tang Y, Eskilsson A, et al. Interleukin-6 primarily produced by non-hematopoietic cells mediates the lipopolysaccharide-induced febrile response. Brain Behav Immun. 2013;33:123–130. doi:10.1016/j.bbi.2013.06.006
25. Hubbard LL, Ballinger MN, Wilke CA, Moore BB. Comparison of conditioning regimens for alveolar macrophage reconstitution and innate immune function post bone marrow transplant. Exp Lung Res. 2008;34(5):263–275. doi:10.1080/01902140802022518
26. Zhou X, Loomis-King H, Gurczynski SJ, et al. Bone marrow transplantation alters lung antigen-presenting cells to promote TH17 response and the development of pneumonitis and fibrosis following gammaherpesvirus infection. Mucosal Immunol. 2016;9(3):610–620. doi:10.1038/mi.2015.85
27. Fan Y, Zhang G, Vong CT, Ye RD. Serum amyloid A3 confers protection against acute lung injury in Pseudomonas aeruginosa-infected mice. Am J Physiol Lung Cell Mol Physiol. 2020;318(2):L314–L322. doi:10.1152/ajplung.00309.2019
28. Percopo CM, Ma M, Brenner TA, et al. Critical adverse impact of IL-6 in acute pneumovirus infection. J Immunol. 2019;202(3):871–882. doi:10.4049/jimmunol.1800927
29. Savale L, Tu L, Rideau D, et al. Impact of interleukin-6 on hypoxia-induced pulmonary hypertension and lung inflammation in mice. Respir Res. 2009;10:6. doi:10.1186/1465-9921-10-6
30. Yadav H, Nolan ME, Bohman JK, et al. Epidemiology of acute respiratory distress syndrome following hematopoietic stem cell transplantation. Crit Care Med. 2016;44(6):1082–1090. doi:10.1097/CCM.0000000000001617
31. Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ, et al. Sepsis: multiple abnormalities, heterogeneous responses, and evolving understanding. Physiol Rev. 2013;93(3):1247–1288. doi:10.1152/physrev.00037.2012
32. Denham W, Norman J. The potential role of therapeutic cytokine manipulation in acute pancreatitis. Surg Clin North Am. 1999;79(4):767–781. doi:10.1016/s0039-6109(05)70042-6
33. Huang L, Zhao X, Qi Y, et al. Sepsis-associated severe interleukin-6 storm in critical coronavirus disease 2019. Cell Mol Immunol. 2020;17(10):1092–1094. doi:10.1038/s41423-020-00522-6
34. Meynaar IA, Droog W, Batstra M, Vreede R, Herbrink P. In critically ill patients, serum procalcitonin is more useful in differentiating between sepsis and SIRS than CRP, Il-6, or LBP. Crit Care Res Pract. 2011;2011:594645. doi:10.1155/2011/594645
35. Burgmeijer EH, Duijkers R, Lutter R, Bonten MJM, Schweitzer VA, Boersma WG. Plasma cytokine profile on admission related to aetiology in community-acquired pneumonia. Clin Respir J. 2019;13(10):605–613. doi:10.1111/crj.13062
36. Siljan WW, Holter JC, Nymo SH, et al. Cytokine responses, microbial aetiology and short-term outcome in community-acquired pneumonia. Eur J Clin Invest. 2018;48(1):e12865. doi:10.1111/eci.12865
37. Doring M, Cabanillas Stanchi KM, Mezger M, et al. Cytokine serum levels during post-transplant adverse events in 61 pediatric patients after hematopoietic stem cell transplantation. BMC Cancer. 2015;15:607. doi:10.1186/s12885-015-1616-z
38. Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108(1):17–41. doi:10.1002/JLB.3COVR0520-272R
39. Gao R, Bhatnagar J, Blau DM, et al. Cytokine and chemokine profiles in lung tissues from fatal cases of 2009 pandemic influenza A (H1N1): role of the host immune response in pathogenesis. Am J Pathol. 2013;183(4):1258–1268. doi:10.1016/j.ajpath.2013.06.023
40. Voiriot G, Razazi K, Amsellem V, et al. Interleukin-6 displays lung anti-inflammatory properties and exerts protective hemodynamic effects in a double-hit murine acute lung injury. Respir Res. 2017;18(1):64. doi:10.1186/s12931-017-0553-6
41. Bhargava R, Janssen W, Altmann C, et al. Intratracheal IL-6 protects against lung inflammation in direct, but not indirect, causes of acute lung injury in mice. PLoS One. 2013;8(5):e61405. doi:10.1371/journal.pone.0061405
42. Yang ML, Wang CT, Yang SJ, et al. IL-6 ameliorates acute lung injury in influenza virus infection. Sci Rep. 2017;7:43829. doi:10.1038/srep43829
43. Dienz O, Rud JG, Eaton SM, et al. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 2012;5(3):258–266. doi:10.1038/mi.2012.2
44. Murphy EA, Davis JM, Brown AS, Carmichael MD, Ghaffar A, Mayer EP. Effect of IL-6 deficiency on susceptibility to HSV-1 respiratory infection and intrinsic macrophage antiviral resistance. J Interferon Cytokine Res. 2008;28(10):589–595. doi:10.1089/jir.2007.0103
45. Ibrahim YF, Moussa RA, Bayoumi AMA, Ahmed AF. Tocilizumab attenuates acute lung and kidney injuries and improves survival in a rat model of sepsis via down-regulation of NF-kappaB/JNK: a possible role of P-glycoprotein. Inflammopharmacology. 2020;28(1):215–230. doi:10.1007/s10787-019-00628-y
46. Investigators R-C, Gordon AC, Mouncey PR, et al. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N Engl J Med. 2021;384(16):1491–1502. doi:10.1056/NEJMoa2100433
47. McGonagle D, Sharif K, O’Regan A, Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev. 2020;19(6):102537. doi:10.1016/j.autrev.2020.102537
48. Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13(12):862–874. doi:10.1038/nri3552
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.