Back to Journals » Infection and Drug Resistance » Volume 10
Performance of VITEK mass spectrometry V3.0 for rapid identification of clinical Aspergillus fumigatus in different culture conditions based on ribosomal proteins
Received 3 August 2017
Accepted for publication 31 October 2017
Published 8 December 2017 Volume 2017:10 Pages 499—506
DOI https://doi.org/10.2147/IDR.S148121
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Longrong Zhou, Yongquan Chen, Yuanhong Xu
Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Anhui, Hefei, People’s Republic of China
Abstract: Fast and accurate discrimination of Aspergillus fumigatus is significant, since misidentification may lead to inappropriate clinical therapy. This study assessed VITEK mass spectrometry (MS) V3.0 for A. fumigatus identification using extracted fungal ribosomal proteins. A total of 52 isolates preliminarily identified as A. fumigatus by traditional morphological methods were inoculated in three different culture media and cultured at two different temperatures. The specific spectral fingerprints of different culture time points (48, 72, 96, and 120 h) were obtained. Of all strains, 88.5% (46/52) were discriminated as A. fumigatus, while the remaining 11.5% (6/52) produced results inconsistent with morphological analysis. Molecular sequencing, as a reference method for species identification, was used to validate the morphological analysis and matrix-assisted laser desorption/ionization time of flight MS. Chi-square tests (Χ2 test, P=0.05) demonstrated that the culture medium and incubation temperature had no effects on identification accuracy; however, identification accuracy of the strains in the 48-h group was lower than that in other groups. In addition, we found that ribosomal proteins extracted from A. fumigatus can be stored in different environments for at least 1 week, with their profiles remaining stable and strain identification results showing no change. This is beneficial for medical institutions with no mass spectrometer at hand. Overall, this study showed the powerful ability of VITEK MS V 3.0 in identifying A. fumigatus.
Keywords: VITEK MS V 3.0, Aspergillus fumigatus, identification, ribosomal protein, spectral fingerprints, fungal, matrix assisted laser desorption ionization-time of flight mass spectrometry, MALDI-TOF MS
Introduction
In recent years, with the overuse and misuse of antibiotics and non-standard use of chemotherapeutic drugs, humans are increasingly susceptible to infection by fungi,1–4 especially Aspergillus fumigatus. Infection by this pathogen results in high morbidity and mortality. For clinical laboratory technicians, rapid identification of A. fumigatus is very significant for patient health. A. fumigatus is a multicellular fungus, composed of hyphae, spores, and mycelial elongation branches, intertwined into groups.5 Its identification in clinical laboratories is still performed by morphological methods,6 histopathology, and serology. However, such methods are cumbersome, relatively difficult to carry out, have long detection periods, and non-ideal sensitivity and specificity. To further improve A. fumigatus identification, sequencing techniques attract increasing attention, not requiring complex identification criteria for the clinical setting or long incubations of filamentous fungi used in conventional phenotypic methods.7 However, long detection cycles and high detection costs restrict the application to a relatively low number of institutions; therefore, it cannot be used as routine detection tools for A. fumigatus.
Fortunately, clinical microbiology laboratories have significantly shortened the identification time of A. fumigatus in recent years using a new technique, VITEK mass spectrometry (MS),8,9 which is matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) based. VITEK MS is a new identification tool that can directly quantify biomarkers (eg, ribosomal proteins) to identify viruses, bacteria, fungi, and other microorganisms,10–13 with high sensitivity, throughput, and speed. Importantly, this technology was used to identify A. fumigatus with quite satisfactory results8,9 and is clinically important for related infections caused by filamentous fungi (mainly A. fumigatus). However, whether culture conditions (eg, culture medium and incubation temperature) and time affect A. fumigatus identification results remains unclear.10,14–22 The current study addressed this issue.
Materials and methods
Fungal strains and culture conditions
The 52 isolates used in this study were clinical specimens from patients hospitalized in the First Affiliated Hospital of Anhui Medical University and Anhui Provincial Hospital (isolated from sputum, urine, or tissue samples) between November 2016 and April 2017. When the same strain was detected more than once in a patient’s specimens, only one of them was considered. The isolates were identified as A. fumigatus by morphological (macroscopic and microscopic) tools preliminarily, as described previously.23 Pure cultures were preserved in 25% (v/v) glycerol at -80°C until use. Escherichia coli ATCC 8739 (gift from Biomerieux, France) was used as a quality control strain. Each clinical isolate was inoculated on three fungal media, including Sabouraud dextrose liquid, Sabouraud dextrose agar (SDA), and Chalet agar (Hangzhou Bai Si Biotechnology Co, Ltd, Hangzhou, People’s Republic of China), and cultured at 28°C and 35°C, respectively.
Sample preparation for mass spectrometry, spectrum generation, and data analysis
Strains cultured for 48, 72, 96, and 120 h were assessed. For isolates grown on solid media (including Sabouraud- and Chalet-agar media), 1 μL of colonies (or ~2 cm in diameter) were scraped with a sterile scalpel (or cotton swabs/disposable plastic inoculation rings) and resuspended in 300 μL of distilled water. After addition of 900 μL of anhydrous ethanol (AR), the samples were centrifuged at 10,000 rpm for 2 min. The resultant supernatant was removed, while the pellet was mixed with 40 μL of 70% formic acid; then, the same volume of acetonitrile was added, followed by centrifugation (10,000 rpm) for 2 min. The supernatant containing the proteins of interest (1 μL) was spotted on the mass spectrometer sample target (Biomerieux) and air-dried naturally. Next, 1 μL of α-hydroxy-4-cyanomethyl cinnamic acid (CHCA, Biomerieux) was used to cover the target plate before the assessment.
The procedures for strains grown in liquid media (Sabouraud liquid media) were as follows. Appropriate amounts of colonies were collected from the Sabouraud liquid medium and placed in 1.5 mL tubes. After high-speed centrifugation (10,000 rpm) for 10 min, the culture medium was removed, and the spores and hyphae were collected and resuspended in 1.2 mL of distilled water. After vortex mixing the sample, high-speed centrifugation (10,000 rpm) was performed for 2 min, discarding the supernatant. Two additional washes with distilled water were performed. The subsequent steps were carried out as described earlier for solid culture media.
Spectra were recorded in the linear positive mode at a laser frequency of 20 Hz within a mass range of 2000–20,000 Da by shooting each spot 108 times. Before analysis, the quality control strain E. coli ATCC8739 (Biomerieux) was used to calibrate the mass spectrometer.
Molecular sequence identification
To obtain accurate and reliable identification results, three genes (ITS, β-tubulin, and calmodulin) were used to characterize target strains. The base sequence of Its1 was 5’-TCCGTAGGTGAACCTGCGG-3’, and the base sequence of Its4 was 5’-TCCTCCGCTTATTGATATGC-3’.24 The following primers were used for β-tubulin: forward (F), 5’-TGACGGGTGATTGGGATCTC-3’; reverse (R), 5’-CGTCCGCTTCTTCCTTGTTT-3’.25 The primers cmd5 (5’-CCGAGTACAAGGAGGCCTTC-3’) and cmd6 (5’-CCGATAGAGGTCATAACGTGG-3’) were used to amplify the calmodulin gene.26 Strains were inoculated on SDA plates and incubated for 2 days at 28°C. Shanghai Biotechnology Company carried out DNA extraction, amplification, and sequencing, based on the above primers. Sequencing results were compared with GenBank by BLAST27 and UniProt Knowledgebase (UniProtKB);28 homology ≥97% and ≥99% reflected appurtenance to the same genus and species, respectively.
Ethical approval
This study was approved by the ethical committee of the First Affiliated Hospital of Anhui Medical University. Written informed consent was obtained from the patients whose samples were used.
Results
(I) A total of 52 A. fumigatus strains identified by morphological methods were also identified by VITEK MS, and the multi-gene detection method was employed to verify morphological data and VITEK MS findings. As shown in Table 1, VITEK MS identification showed 11.5% (6/52) inconsistent findings with morphological results, while 88.5% (46/52) were consistent. Most importantly, the VITEK MS technique was consistent with the molecular method.
(II) VITEK MS identified A. fumigatus in different culture conditions (Table 2).
For strains cultured in different culture media, the SPSS software revealed c2=1.660 (P=0.436), indicating that the culture medium had no effect on the discrimination rate of A. fumigatus. When considering only temperatures, c2=1.029 (P=0.296), indicating no effect of incubation temperature on the discrimination rate.
However, for strains grown at different time points, c2=18.677 (P=0.000), suggesting that the MALDI-TOF-MS technique did not have exactly the same capacity for identifying A. fumigatus cultured at different time points. The 72 h, 96 h, and 120 h groups showed statistically significant differences compared with the 48-h group, which displayed the lowest discrimination rate.
The remaining six strains were identified as A. flavus or A. oryzae by VITEK MS, and these results were consistent with molecular findings.
(III) Identification of proteins stored at different times in various environments. In this study, we observed an interesting phenomenon: the proteins extracted by formic acid–acetonitrile were very stable in in vitro conditions and could be stored in different environments. The 72-h group cultured at 35°C (containing carbon dioxide) was assessed, selecting 15 A. fumigatus strains randomly; the extracted proteins were stored at 4°C, -20°C and -80°C for 48 h, 72 h, 120 h and 168 h. Identification results are listed in Table 3.
  | Table 3 Identification results for proteins stored at different temperatures for various time periods |
The above-mentioned results showed that the extracted proteins could be stored at 4°C, -20°C, and -80°C for at least 168 h (1 week), with identification results almost not affected by storage time or temperature; in addition, mass spectra at different time periods also showed no significant changes (Figures 1–3).
  | Figure 1 Proteins stored at 4°C. Note: The 0 h, 48 h, 72 h, 120 h, and 168 h groups, respectively, are presented from bottom to top. |
  | Figure 2 Proteins stored at -20°C. Note: The 0 h, 48 h, 72 h, 120 h, and 168 h groups, respectively, are presented from bottom to top. |
  | Figure 3 Proteins stored at -80°C. Note: The 0 h, 48 h, 72 h, 120 h, and 168 h groups, respectively, are presented from bottom to top. |
Discussion
This study demonstrated that for A. fumigatus grown either in liquid or on solid culture media, identification accuracy was stable after 72 h of culture and did not change with incubation time or culture conditions. These results addressed a question by Posteraro et al10 whether mold colonies cultured on solid media might be more heterogeneous than those grown in liquid media. Although A. fumigatus organisms have different components (including spores, fruiting bodies, surface hyphae, and basal mycelia) in different growth periods,9 identification results remained stable using VITEK MS after 72 h of culture and were not affected by product composition of the strain. These findings suggest that regardless of culture environment, A. fumigatus identification remains stable. Compared with 48-h cultures, the strains were more favorable for analysis after 72-h of incubation. The reason might be that a few strains cultured for 48 h have smaller colony products than those for 72 h, resulting in some ribosomal proteins not being extracted, which directly affects mass spectrometry data. Although identification after 72 h is more convenient, there is no significant difference in time compared with the traditional method; however, this technique is more accurate, and the patient can receive symptomatic treatment as soon as possible, with shortened hospitalization time and reduced treatment costs. VITEK MS is a relatively fast identification tool for A. fumigatus.
Importantly, in this study, we found that proteins extracted by formic acid–acetonitrile could be stored for at least 168 h (1 week) at 4°C, -20°C, or -80°C. Indeed, the proteins extracted from A. fumigatus strains were ribosomal proteins, which are typical housekeeping molecules abundantly present in microbial cells. Identification results did not vary significantly with storage time prolongation, suggesting that ribosomal proteins are very stable. There are ~78 ribosomal subunit proteins in eukaryotes. Nakamura et al selected and verified 26 kinds of ribosomal subunit proteins (RSPs) whose molecular weights were under 16000 Da that could be used as biomarkers for A. fumigatus strains.19,29 In conjunction with his study, we observed 20 kinds of RSPs that could be used as biomarkers when VITEK MS was used to identify our strains. Table 4 shows the RSP types selected in this study. Each strain expressed all 20 RSPs. Although some RSPs have the same m/z ratio in other aspergilli, we compared the RSPs of common aspergilli, such as A. fumigatus, A. flavus, A. oryzae, and A. niger. Among the 20 RSPs, A. flavus and A. oryzae could not be differentiated, because their genes are essentially the same. Assessing A. fumigatus and A. flavus/A. oryzae, we found that they had exactly the same L39, while A. niger and A. flavus/A. oryzae had the same L43 and S26. However, this did not affect identification results, because each strain has a set of unique ribosomal proteins, with a unique mass spectrum fingerprint; even if individual ribosomal protein identical in some strains, they have no significant effect on mass spectrum fingerprints.
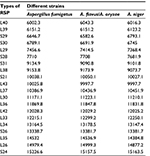  | Table 4 Types of RSP used in this study Abbreviation: RSP, ribosomal subunit protein. |
For clinical laboratory technicians, providing accurate results of the patient’s isolates as soon as possible is of outmost importance. The MALDI-TOF MS technology has been developed rapidly and successfully applied in routine identification of clinical microorganisms. The stability of ribosomal proteins is beneficial to many medical institutions. As mass spectrometers are still expensive, only a few large medical institutions can afford them; therefore, those without mass spectrometer can extract ribosomal proteins from clinical isolates and send to relevant agencies for mass spectrometry identification. Many laboratories used MALDI-TOF MS to identify common isolates quickly and accurately. However, compared with bacteria, the MALDI-TOF MS technology is seldom used to identify fungi, and especially rare for the analysis of Aspergillus, Penicillium, and other filamentous fungi. This can be explained by many reasons. The first and most important point is that the cell wall of filamentous fungi is very thick and hard, requiring a special treatment for ribosomal protein extraction; although the formic acid–acetonitrile extraction method is quite useful, it remains complex and time-consuming for daily clinical application, particularly for strains grown in liquid media or positive blood cultures, since culture media and other nutrients may interfere with mass spectrometry analysis; therefore, colonies must be washed as much as possible before the test.20 Second, the current database covers a limited number of strains and does not allow the detection of all strains; for rare strains, the identification rates are generally low, and sometimes they are not correctly identified. For example, the current method cannot be used to detect Mucor, which is not included in the library.
Conclusion
Overall, MALDI-TOF MS can be used to identify A. fumigatus quickly and accurately. Moreover, this technology has many advantages, such as stability, high-throughput potential, and relatively low cost. In clinical microbiological laboratories, the MALDI-TOF MS technology has brought a revolutionary breakthrough for A. fumigatus identification. However, its shortcomings still require studies for further optimization, after which MALDI-TOF MS in the near future is expected to replace the current identification methods.
Acknowledgments
The materials used in this study were provided by the First Affiliated Hospital of Anhui Medical University and Anhui Provincial Hospital, to which we are grateful. We thank Mrs Ying Huang, Mr Zhongxin Wang, and Mr Jilu Shen, who provided and managed the VITEK MS instrument. This work was supported by the National Natural Science Foundation of China (81171606).
Disclosure
The authors report no conflicts of interest in this work.
References
Suzuki K, Nakase K, Kyo T, et al. Fatal Trichosporon fungemia in patients with hematologic malignancies. Eur J Haematol. 2010;84(5):441–447. | ||
Chitasombat MN, Kofteridis DP, Jiang Y, Tarrand J, Lewis RE, Kontoyiannis DP. Rare opportunistic (non-Candida, non-Cryptococcus) yeast bloodstream infections in patients with cancer. J Infect. 2012;64(1):68–75. | ||
Nucci M, Marr KA, Vehreschild MJ, et al. Improvement in the outcome of invasive fusariosis in the last decade. Clin Microbiol Infect. 2014;20(6):580–585. | ||
Pagano L, Akova M, Dimopoulos G, Herbrecht R, Drgona L, Blijlevens N. Risk assessment and prognostic factors for mould-related diseases in immunocom-promised patients. J Antimicrob Chemother. 2011;66(suppl 1):i5–i14. | ||
Ni Y, Shang H. Clinical Microbiological Examination. 5th ed. Beijing: People’s Health Publishing House; 2012. | ||
Larone DH. Medically Important Fungi: A Guide to Identification. 5th ed. Washington, DC: ASM Press; 2011. | ||
Clarridge JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17(4):840–862. | ||
Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26(3):547–603. | ||
Sanguinetti M, Posteraro B. MALDI-TOF mass spectrometry: any use for Aspergilli? Mycopathologia. 2014;178(5–6):417–426. | ||
Posteraro B, De Carolis E, Vella A, Sanguinetti M. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev Proteomics. 2013;10(2):151–164. | ||
Kliem M, Sauer S. The essence on mass spectrometry based microbial diagnostics. Curr Opin Microbiol. 2012;15(3):397–402. | ||
Murray PR. What is new in clinical microbiology-microbial identification by MALDI-TOF mass spectrometry: a paper from the 2011 William Beaumont Hospital Symposium on molecular pathology. J Mol Diagn. 2012;14(5):419–423. | ||
Bille E, Dauphin B, Leto J, et al. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin Microbiol Infect. 2012;18(11):1117–1125. | ||
Cassagne C, Ranque S, Normand AC, et al. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One. 2011;6(12):e28425. | ||
Coulibaly O, Marinach-Patrice C, Cassagne C, Piarroux R, Mazier D, Ranque S. Pseudallescheria/Scedosporium complex species identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Med Mycol. 2011;49(6):621–626. | ||
Marinach-Patrice C, Lethuillier A, Marly A, et al. Use of mass spectrometry to identify clinical Fusarium isolates. Clin Microbiol Infect. 2009;15(7):634–642. | ||
Del Chierico F, Masotti A, Onori M, et al. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. Proteomics. 2012;75(11):3314–3330. | ||
Li Yan YE, Ling GUO, Yan Ning MA, et al. Analysis of the clinical application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry technology in the rapid identification of pathogenic fungi. Zhonghua Yi Yuan Gan Ran Xue Za Zhi. 2015;25(15):3361–3363. | ||
Nakamura S, Sato H, Tanaka R, Kusuya Y, Takahashi H, Yaguchi T. Ribosomal subunit protein typing using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) for the identification and discrimination of Aspergillus species. BMC Microbiol. 2017;17(1):100. | ||
Bader O. MALDI-TOF MS based species identification and typing approaches in medical mycology. Proteomics. 2013;13(5):788–799. | ||
Anderson NW, Buchan BW, Riebe KM, Parsons LN, Gnacinski S, Ledeboer NA. Effects of solid-media type on routine identification of bacterial isolates by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50(3):1008–1013. | ||
Yuexia Z, Chengjin H, Yingjian C. MALDI-TOF MS microbiological identification database application research. J Clin Lab Med. 2016;34(6):447–450. | ||
de Hoog GS, Guarro J, Gene J, Figueras MJ. Atlas of Clinical Fungi. 2nd ed. Utrecht, NL: Centraalbureau voor Schimmelcultures; 2000. | ||
Peng-hao GUO, Xiu-li LIU, Ying-peng CUI, et al. The value of universal fungal primers Its1 and Its4 in the clinical identification of filamentous fungi. Chin J Microecol. 2013;25(8):922–924. | ||
Serrano R, Gusmão L, Amorim A, Araujo R. Rapid identification of Aspergillus fumigatus within the section Fumigati. BMC Microbiol. 2011;11:82. | ||
Hong S-B, Go S-J, Shin H-D, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97(6):1316–1329. | ||
BLAST Database [homepage on the Internet]. National Institutes of Health. Available from: http://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed November 30, 2017. | ||
UniProt Database [homepage on the Internet]. UniProt Consortium. Available from: http://www.uniprot.org/. Accessed November 30, 2017. | ||
Nakamura S, Sato H, Tanaka R, Yaguchi T. Verification of ribosomal proteins of Aspergillus fumigatus for use as biomarkers in MALDI–TOF MS identification. Mass Spectrom (Tokyo). 2016;5(1):A0049. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


