Back to Journals » International Journal of General Medicine » Volume 14
PER1 as a Tumor Suppressor Attenuated in the Malignant Phenotypes of Breast Cancer Cells
Authors Liu Y, Hao J, Yuan G, Wei M, Bu Y, Jin T, Ma L
Received 10 July 2021
Accepted for publication 29 September 2021
Published 22 October 2021 Volume 2021:14 Pages 7077—7087
DOI https://doi.org/10.2147/IJGM.S328184
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yinfeng Liu,1,2 Jun Hao,3 Guanli Yuan,4 Mengyu Wei,1 Yuhui Bu,1 Tingting Jin,3 Li Ma1
1Department of Breast Disease Center, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, 050000, People’s Republic of China; 2Department of Breast Surgery, The First Hospital of Qinhuangdao, Qinhuangdao, Hebei, 066000, People’s Republic of China; 3Center of Metabolic Diseases and Cancer Research, Institute of Medical and Health Science of Hebei Medical University, Shijiazhuang, Hebei, 050000, People’s Republic of China; 4Department of Respiratory and Critical Care Medicine, The First Hospital of Qinhuangdao, Qinhuangdao, Hebei, 066000, People’s Republic of China
Correspondence: Li Ma
Department of Breast Disease Center, The Fourth Hospital of Hebei Medical University, No. 169, Tianshan Street, Shijiazhuang, Hebei, 050000, People’s Republic of China
Tel +86 -0311-66696348
Email [email protected]
Background: Circadian clock genes play a crucial role in physiological and pathological processes, and their aberrant expressions were involved in various human cancers. The objective of this study was to investigate the expression level of Period circadian regulator 1 (PER1), an important circadian clock gene, and its biological roles in the development and progression of breast cancer.
Methods: The expression level of PER1 in breast cancer samples was analyzed using the Oncomine database, and the correlation between PER1 expression and clinicopathologic parameters was assessed by bc-GenExMiner v4.5. In addition, Kaplan–Meier plotter database was used to determine the prognostic significance of PER1 expression for breast cancer patients. The expressions of PER1 in breast cancer tissues and cells were validated by Western blot. The loss-or-gain assay of PER1 was conducted to investigate the effects of its expression on cell proliferation, migration and invasion of breast cancer. The relationship between PER1 expression and epigenetic modifications was further explored by Western blot.
Results: The results of the bioinformatics analysis revealed that the expression level of PER1 was markedly reduced in breast cancer tissues (P< 0.001), and patients with high expression of PER1 had a better overall survival (HR:0.78, 95% CI:0.63– 0.97, P=0.026) and recurrence-free survival (HR:0.83, 95% CI:0.75– 0.93, P=0.001) than those with low expression. The assay of gene loss-or-gain indicated that downregulation of PER1 expression markedly promoted cell proliferation, migration and invasion (P< 0.05), whereas these malignant phenotypes of breast cancer cells were inhibited by PER1 overexpression (P< 0.05). Further studies showed that trichostatin A (TSA), a histone deacetylase inhibitor, induced the expression of PER1 protein in breast cancer cells (P< 0.05).
Conclusion: PER1 functions as a tumor suppressor in the development and progression of breast cancer, and its expression silencing might be regulated by epigenetic modifications.
Keywords: breast cancer, circadian clock gene, Period circadian regulator 1, PER1, tumor suppressor
Introduction
Breast cancer is one of the most common malignancies in women, with a high mortality rate around the world.1,2 Although multidisciplinary therapeutic strategies, including surgical resection, chemoradiotherapy, endocrine and anti-HER2 therapy, have made substantial progress over the past decade, there are still a considerable proportion of breast cancer patients who develop loco-regional recurrence and distant metastasis and die of this malignancy.3,4 Thus, identifying novel and effective biomarkers is helpful for early diagnosis, prognostic assessment and individualized treatment of breast cancer patients.
It has been well established that major cellular activities of human beings, such as growth, metabolism and hormone secretion, are controlled by circadian rhythm.5 As an important circadian clock gene, period circadian regulator 1 (PER1) was reported to be mainly responsible for circadian rhythms and coordinated the basic activities of life.6,7 Recently, emerging evidence has shown that PER1 is abnormally expressed in several human cancers and involved in tumorigenesis and progression.8–10 Several studies reported that the expression level of PER1 was markedly downregulated in oral squamous cell carcinoma (OSCC) and cholangiocarcinoma tissues,11,12 and PER1 overexpression suppressed proliferation, migration and invasion and promoted autophagy and apoptosis of cancer cells.11,12 These findings suggested that PER1 might function as a tumor suppressor in the development and progression of human cancers. However, whether PER1 played a similar role in the pathogenesis of breast cancer remains unclear. Microarray and RNA-sequencing technologies have made comprehensive analysis of specific genes possible. In the present study, we mainly focused on the biological roles of PER1 in the development and progression of breast cancer. For this purpose, the expression level of PER1 and its relationship to clinicopathological features and prognosis of breast cancer patients were analyzed using the public database. Next, a loss-or-gain function assay was conducted to investigate the effects of PER1 expression on cell proliferation, migration and invasion of breast cancer. We believed that these findings provided novel insights into the pathogenesis of breast cancer and therapeutic targets.
Materials and Methods
Bioinformatics Analysis
The expression levels of PER1 mRNA in human cancers were analyzed using the Oncomine microarray database (http://www.oncomine.com).13 The correlation between the expression of PER1 and the clinicopathologic parameters of breast cancer was evaluated by Breast cancer gene expression miner (bc-GenExMiner) v4.5, which was an online mining tool for transcriptome data of breast cancer (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php).14 To further investigate the prognostic significance of PER1 expression for breast cancer patients, Kaplan–Meier plotter (https://kmplot.com/analysis/),15 an online database involving gene expression and survival data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), was used. The primary outcomes of survival analysis were overall survival (OS) and recurrence-free survival (RFS). All patients were divided into high and low-expression group based on the median of PER1 expression. The Kaplan–Meier curves of the two groups were shown, and the results were expressed as hazard ratio (HR) and its 95% confidence intervals (CI).
Cell Culture and Tissue Samples
Five human breast cancer cell lines (including MCF-7, MDA-MB-453, MDA-MB-231, SKBR3 and BT-549) and normal breast epithelial cell-line MCF-10A were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). All the cell lines were cultured with DMEM (Invitrogen, USA) containing 10% fetal bovine serum (GIBCO, USA) and penicillin–streptomycin (100 U/mL), and were maintained at 37°C with 5% CO2.
Four paired breast cancer samples were collected from patients who were treated with surgical resection, and no patients underwent neoadjuvant chemoradiotherapy and anti-HER2 therapy before surgery. The written informed consent was signed by each patient in order to notify the applying purpose of tissue sample. The study was approved by the medical ethics committee of the Fourth Hospital of Hebei Medical University (No. 2020KY113), and all experiments were conducted in accordance with the Declaration of Helsinki.
Plasmid Construction and Transfection
The short hairpin RNA (shRNA) targeted to PER1 (5ʹ-CCAGCACCACTAAGCGTAAAT-3ʹ) were designed and synthesized, and the specific sequence was cloned into the pGenesil-1 vector (pGenesil1-PER1) (Wuhan Genesil Biotechnology, China). The pGenesil-1 vector with the expression of green fluorescent protein (GFP) was used as negative control (pGenesil1-KB). On the other hand, pCDNA3.1 vector expressing PER1 (pCDNA3.1-PER1) was used for transfecting with breast cancer cells. To establish a stable cell line with PER1 overexpression, we also used the recombinant lentiviral plasmids with PER1 sequence (Lenti-blast-PER1) to transfect into MDA-MB-231 cells according to the manufacturer’s protocol. The cells were inoculated in a six-well plate at a density of 5*105 cells/well. After transfection for 24 h, cells were further screened by treatment with blasticidin for 2 weeks. Cells in the control group were transfected with empty lentiviral plasmid (Lenti-blast-KB). The efficiency of transfection was assessed under a fluorescence microscope (NIKON, Japan).
Western Blotting
Briefly, total proteins from breast cancer tissues and cells were isolated using radio-immunoprecipitation assay (RIPA) lysis buffer with PMSF and protease inhibitor. BCA Protein Assay Kit was used to quantify the concentration of total proteins, and then they were separated by 10–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto polyvinylidene difluoride (PVDF) membranes. Next, PVDF membranes were blocked by 5% non-fat milk and were incubated with primary antibodies against PER1 (1:1000, GTX128974, GENETEX, USA) and β-actin (1:10,000, AC026, Abclonnal, China) at 4°C overnight and secondary antibody (1:5000, SA00001-2, Proteintech, China) for 2 h at room temperature. The protein bands were washed three times by PBS and then were visualized by enhanced chemiluminescence (ECL).
Colony Formation Assay
Breast cancer cells were collected during the logarithmic growth phase and were seeded into six-well plates with 700 cells per well after transfection. Then, cells were fixed with methanol and stained with 0.1% crystal violet after 14 days. The amount of cloned cells was counted under a light microscope (Olympus, Japan), and the rate of colony formation was calculated as the ratio of colonies to inoculated cells.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
EdU assay was conducted to further evaluate the effects of PER1 expression on the proliferation of breast cancer cells. In brief, transfected cells were seeded into 24-well plates at a density of 5000 cells/well. After that, cells were incubated in medium with EdU solution (RiboBio, China) for 2 h, and then were fixed with 4% paraformaldehyde and stained by DAPI. The percentage of EdU-positive cells was counted under a fluorescence microscope (Olympus, Japan).
Cell Migration and Transwell Invasion
The migration and invasion ability of breast cancer cells in vitro was evaluated in transwell chamber. Briefly, the transfected cells were suspended in a serum-free medium and were seeded into the upper transwell chambers with 5×104 cells per well.
In contrast, the lower transwell chambers were filled with 600 μL cell medium containing 10% fetal bovine serum. For the Transwell invasion assay, the upper chambers were additionally coated with 30 μg Matrigel (Corning, USA), and other procedures were performed the same as the cell migration assay. After 24 h incubation, the infiltrating cells into the bottom surface were fixed with 4% formaldehyde and stained with 0.1% crystal violet. Then, the amount of these cells were counted under 5 random fields using a laser confocal microscope.
Wound Healing Assay
For wound healing assay, breast cancer cells were transfected and cultured in six-well plates (5 × 104 cells per well) for 24 h. Next, these cells were scratched by a 200μL pipette tip and washed three times with PBS. The images of the migrating cells were captured at 0 and 24 h under a light microscope (Olympus, Japan), respectively.
Treatment of Epigenetic Modifications and Proteasome Inhibition
Breast cancer cells were treated with DNA methyltransferase inhibitor 5-Aza-2ʹ-deoxycytidine (5-AzaC) (Sigma-Aldrich, USA), histone deacetylase inhibitor trichostatin A (TSA) (Sigma-Aldrich, USA), MA2 (Shanghai Institiute of Materia Medica, China) or proteasome inhibitor MG132 (Sigma-Aldrich, USA). After that, the cells were harvested to detect the expression of PER1 using Western blotting.
Immunofluorescence Assay
The expression of PER1 in breast cancer cells was further detected by immunofluorescence staining. Briefly, cells were treated with TSA and then were fixed with 4% paraformaldehyde for 15 min, permeabilized by 0.5% Triton X-100 and blocked with 5% Bovine serum for 1 h at room temperature. Subsequently, cells were incubated with rabbit polyclonal anti-PER1 antibody (1:100, 13463-1-AP, Proteintech, China) at 4°C overnight. After washing with PBS for three times, the cells were incubated with fluorescent-labeled secondary antibody (1:200, RS23320, Immunoway, USA) in the dark for 1 h. The nuclei were stained with DAPI, and the images were photographed under a fluorescent microscope (Olympus, Japan).
Statistical Analysis
The differential expression between breast cancer tissues and adjacent normal tissues was analyzed by Wilcoxon test. Student’s t-test and/or Kruskal-Wallis H-test were used to evaluate the relationship between the expression of PER1 and clinicopathologic features of breast cancer. The survival difference between high and low PER1 expression groups was compared by the Log rank test. Univariate and multivariate Cox regression analysis was conducted to determine independent prognostic factors for breast cancer patients. All experiments were conducted at least 3 times, and the data were expressed as mean ± standard deviation (SD). The comparison between the study groups was carried out using Student’s t-test. All data were analyzed using the SPSS 21.0 version of the software package (IBM Corp, Chicago, USA). A p-value <0.05 was considered to be a statistically significant difference.
Results
PER1 Expression Was Downregulated in Breast Cancer Tissues
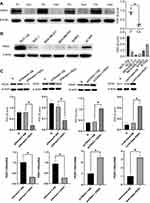
Using the Oncomine online database, we first detected the alteration of PER1 mRNA expression in various human cancers (Figure 1A). We found that the expression level of PER1 was significantly downregulated in most human cancers (Figure 1B). Moreover, the expression of PER1 mRNA was significantly lower in breast cancer tissues than in adjacent non-cancer tissues (P<0.001) (Figure 1C). Similar findings were observed in paired breast cancer samples (P<0.001) (Figure 1D). Next, we investigated the correlations between PER1 mRNA expression and clinicopathological features of breast cancer using the bc-GenExMiner v4.5 database. Our results indicated that low expression of PER1 was significantly related to estrogen receptor (ER)-negative status (P<0.001), progesterone receptor (PR)-negative status (P<0.001), human epidermal growth factor receptor 2 (HER2)-positive status (P<0.001), basal-like status (P<0.001) and triple-negative status (P<0.001) of breast cancer samples (Figure 2). Of note, Luminal B type had the lowest expression level of PER1 mRNA compared with other molecular subtypes of breast cancer (P<0.001). Based on the Nottingham prognostic index (NPI), we found that the expression of PER1 was markedly down-regulated in NPI grade 2 and grade 3 (P<0.001). In terms of Scarff Bloom Richardson (SBR) grading system, the results showed that low expression of PER1 was significantly associated with higher SBR grade (P<0.001) (Figure 2).
 |
Figure 2 The correlations between PER1 mRNA expression and clinicopathological features of breast cancer were analyzed by the bc-GenExMiner v4.5 database. |
Low Expression of PER1 Was Associated with Poor Survival in Breast Cancer Patients
Based on the above findings, we further evaluated the prognostic significance of PER1 mRNA expression for breast cancer patients using the Kaplan–Meier plotter database. As shown in Figure 3A, Kaplan–Meier curves revealed that breast cancer patients with high PER1 expression had a better overall survival (OS) [HR:0.78, 95% CI:0.63–0.97, P=0.026] and recurrence-free survival (RFS) [HR:0.83, 95% CI:0.75–0.93, P=0.001] than those with low PER1 expression (Figure 3A). In addition, subgroup analysis based on different clinicopathological features further revealed the prognostic value of PER1 for breast cancer patients (Figure 3B).
PER1 Expression Regulated Cell Proliferation, Migration and Invasion of Breast Cancer
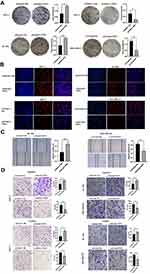
To further validate the results of the bioinformatic analysis, we also detected the expression level of PER1 in four paired samples of breast cancer using Western blot. The results showed that the expression of PER1 was markedly lower in breast cancer tissues than that in adjacent normal tissues (P<0.05) (Figure 4A). On the other hand, PER1 expression was significantly decreased in five breast cancer cell lines (MCF-7, MDA-MB-231, MDA-MB-453, SKBR3 and BT-549) compared with normal breast epithelial cell-line MCF-10A (P<0.05) (Figure 4B).
Among the five breast cancer cell lines, MDA-MB-231 had the lowest expression level, BT-549 had the highest expression level, and MCF-7 had an intermediate expression. Thus, three cell lines were further used to conduct the function assay of loss-or-gain expression. Figure 4C shows the transfected efficiency of PER1 overexpression and/or knockdown in breast cancer cells. The expression levels of PER1 in MCF-7 and BT-549 cells were significantly down-regulated by transfecting with pGenesil1-PER1 (P<0.05). In contrast, the plasmids with pCDNA3.1-PER1 or Lenti-blast-PER1 significantly up-regulated the expression of PER1 in MCF-7 or MDA-MB-231 cells (P<0.05) (Figure 4C).
The results of the colony formation assay demonstrated that PER1 knockdown markedly increased the number of cloned cells (P<0.05), while PER1 overexpression inhibited breast cancer cell growth compared with the control group (P<0.05) (Figure 5A). On the other hand, EDU assay also indicated that PER1 knockdown evidently promoted the cell proliferation of breast cancer (P<0.05). By comparison, the proliferative ability of MCF-7 and MDA-MB-231 cells could be suppressed by the upregulation of PER1 expression (P<0.05) (Figure 5B).
We next explored the effects of PER1 expression on the cell migration and invasion of breast cancer. As shown in Figure 5C, the scratched area was evidently reduced by PER1 knockdown (P<0.05), but opposite findings were observed in PER1-overexpressed cells (P<0.05) (Figure 5C). Similarly, pGenesil1-PER1 group had a larger number of migratory and invasive MCF-7 and BT-549 cells than pGenesil1-KB group (P<0.05). In contrast, PER1 overexpression had an inhibitory effect on the migration and invasion ability of breast cancer cells (P<0.05) (Figure 5D).
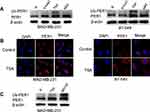
TSA and MA2 Induced the Expression of PER1 in Breast Cancer Cells
To further determine whether the expression of PER1 was regulated by epigenetic modifications, MDA-MB-231 and BT-549 cells were treated with 5-Aza-2ʹ-deoxycytidine (5-AzaC), trichostatin A (TSA) and MA2, respectively. The results showed that the expression levels of PER1 protein (130 KD) and ubiquitinated PER1 protein (180 KD) in breast cancer cells were increased by TSA and MA2 (P<0.05) (Figure 6A). Moreover, TSA markedly enhanced the fluorescence intensity of PER1 expression in both MDA-MB-231 and BT-549 cells (Figure 6B). However, there was no significant increase in PER1 expression for breast cancer cells treated with 5-AzaC. Interestingly, proteasome inhibitor MG132 further increased the expression of ubiquitinated PER1 protein induced by TSA in MDA-MB-231 cells (Figure 6C). These findings suggested that the increased expression of PER1 protein induced by TSA might be mediated by inhibiting degradation of the proteasome.
Discussion
Previous studies have reported that circadian clock genes are involved in physiological and pathological processes and play a crucial role in human health.16–18 Recently, emerging evidence has shown that aberrant expression of PER1, a known clock gene, is associated with carcinogenesis and progression of various human cancers.9,10,12 Yang et al reported that PER1 expression was markedly downregulated in OSCC tissues in comparison to adjacent normal tissues, and its low expression was significantly related to lymph node metastasis, TNM stage and poor survival of OSCC patients.11 In the present study, we mainly focused on the role of PER1 expression in the development and progression of breast cancer. Consistent with previous findings, this study demonstrated that the expression level of PER1 in breast cancer tissues was lower than that in adjacent noncancerous tissue. By conducting bioinformatics analysis, we found that there were significant relationships between low expression of PER1 and unfavorable biological features of breast cancer such as triple-negative status and high grade of NPI and SBR. Kaplan–Meier curves showed that low expression of PER1 had a negative impact on the prognosis of breast cancer patients. These findings suggested that dysregulation of PER1 expression might contribute to tumorigenesis and progression of breast cancer.
To further validate this hypothesis, the biological function of PER1 in the development and progression of breast cancer was investigated. The results demonstrated that downregulation of PER1 expression markedly promoted the proliferation, migration and invasion of breast cancer cells, whereas these malignant phenotypes were inhibited by PER1 overexpression. Our findings supported that PER1 might play a role as a tumor suppressor in the pathogenesis of breast cancer. Similarly, a few previous studies reported the inhibitory effects of PER1 expression on carcinogenesis and tumor progression, and the effects were mainly dependent on the regulation of cell cycle.12,19,20 The study of Yang et al revealed that the expression levels of CyclinD1 and CyclinE were markedly decreased by PER1 overexpression, inhibiting cell proliferation and promoting autophagy and apoptosis of breast cancer.11 Fu et al reported that PER1 mediated S phase arrest and suppressed cell proliferation of OSCC by regulating cyclin- and CDK-related proteins such as CDC25, CDK1, P53 and P21.21
The silencing of circadian-clock genes might be regulated by epigenetic modifications during carcinogenesis. A previous study reported that DNA methyltransferase inhibitor 5-AzaC and histone deacetylase inhibitor TSA induced the expression of PER3 mRNA in myeloid leukemia cells.22 Recently, Hernández-Rosas et al have shown that the expression of PER1 and PER2 in human gastric cancer cells was increased by TSA or sodium butyrate (NaB) in a dose-dependent manner.23 These findings suggested that the epigenetic changes might result in transcriptional silencing of circadian-clock genes. In our study, the expression of PER1 in breast cancer cells was reversed by TSA and MA2, but there was no response to 5-AzaC. Therefore, histone acetylation at the PER1 promoter of breast cancer cells might be involved in the silencing of this tumor suppressor. However, further studies are warranted to fully understand the role of epigenetic modifications in transcriptional regulation of PER1.
Conclusions
In conclusion, the expression level of PER1 was markedly reduced in breast cancer tissues and cells, and its low expression was significantly correlated with specific biological characteristics and poor prognosis of breast cancer. The upregulation of PER1 expression promoted cell proliferation, migration and invasion, suggesting that it functions as a tumor suppressor in the development and progression of breast cancer. Epigenetic modifications might play a key role in the silencing of PER1 expression during carcinogenesis. These findings could provide a potential target for anti-cancer therapy.
Data Sharing Statement
All data generated or analyzed during this study are available from the corresponding author upon reasonable request.
Acknowledgment
Thanks to Professor Sun from Shanghai Institute of Materia Medica for providing us with MA2.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was supported by Basic Research Programs of Natural Science Foundation of Hebei Province (H2020206367 and H2021206157).
Disclosure
The authors declare that they have no conflict of interest.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
2. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451.
3. Salvo EM, Ramirez AO, Cueto J, et al. Risk of recurrence among patients with HR-positive, HER2-negative, early breast cancer receiving adjuvant endocrine therapy: a systematic review and meta-analysis. Breast. 2021;57:5–17. doi:10.1016/j.breast.2021.02.009
4. Riggio AI, Varley KE, Welm AL. The lingering mysteries of metastatic recurrence in breast cancer. Br J Cancer. 2021;124(1):13–26. doi:10.1038/s41416-020-01161-4
5. Granda TG, Liu XH, Smaaland R, et al. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005;19(2):304–306. doi:10.1096/fj.04-2665fje
6. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22(3):375–382. doi:10.1016/j.molcel.2006.03.038
7. Sun Q, Yang Y, Wang Z, et al. PER1 interaction with GPX1 regulates metabolic homeostasis under oxidative stress. Redox Biol. 2020;37:101694. doi:10.1016/j.redox.2020.101694
8. Wang Z, Wang H, Wang Z, et al. Associated analysis of PER1/TUBB2B with endometrial cancer development caused by circadian rhythm disorders. Med Oncol. 2020;37(10):90. doi:10.1007/s12032-020-01415-4
9. Gong X, Tang H, Yang K. PER1 suppresses glycolysis and cell proliferation in oral squamous cell carcinoma via the PER1/RACK1/PI3K signaling complex. Cell Death Dis. 2021;12(3):276. doi:10.1038/s41419-021-03563-5
10. Mostafaie N, Kállay E, Sauerzapf E, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48(7):642–647. doi:10.1002/mc.20510
11. Yang G, Yang Y, Tang H, Yang K. Loss of the clock gene Per1 promotes oral squamous cell carcinoma progression via the AKT/mTOR pathway. Cancer Sci. 2020;111(5):1542–1554. doi:10.1111/cas.14362
12. Han Y, Meng F, Venter J, et al. miR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. J Hepatol. 2016;64(6):1295–1304. doi:10.1016/j.jhep.2016.02.024
13. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi:10.1016/S1476-5586(04)80047-2
14. Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131(3):765–775. doi:10.1007/s10549-011-1457-7
15. Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi:10.1007/s10549-009-0674-9
16. Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi:10.1038/nrg2430
17. Greene MW. Circadian rhythms and tumor growth. Cancer Lett. 2012;318(2):115–123. doi:10.1016/j.canlet.2012.01.001
18. de Almeida Chuffa LG, Seiva FR, Cucielo MS, Silveira HS, Reiter RJ, Lupi LA. Clock genes and the role of melatonin in cancer cells: an overview. Melatonin Res. 2019;2(2):133–157. doi:10.32794/mr11250026
19. Yang X, Wood PA, Ansell CM, et al. The circadian clock gene Per1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiol Int. 2009;26(7):1323–1339. doi:10.3109/07420520903431301
20. Sato F, Nagata C, Liu Y, et al. PERIOD1 is an anti-apoptotic factor in human pancreatic and hepatic cancer cells. J Biochem. 2009;146(6):833–838. doi:10.1093/jb/mvp126
21. Fu XJ, Li HX, Yang K, Chen D, Tang H. The important tumor suppressor role of PER1 in regulating the cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma cells. Onco Targets Ther. 2016;9:2237–2245.
22. Yang MY, Chang JG, Lin PM, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97(12):1298–1307. doi:10.1111/j.1349-7006.2006.00331.x
23. Hernández-Rosas F, Hernández-Oliveras A, Flores-Peredo L, et al. Histone deacetylase inhibitors induce the expression of tumor suppressor genes Per1 and Per2 in human gastric cancer cells. Oncol Lett. 2018;16(2):1981–1990.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.