Back to Archived Journals » Journal of Receptor, Ligand and Channel Research » Volume 8
Pentameric viral ion channels: from structure to function
Authors Surya W, Li Y, Torres J
Received 2 September 2014
Accepted for publication 16 October 2014
Published 22 December 2014 Volume 2015:8 Pages 9—18
DOI https://doi.org/10.2147/JRLCR.S36064
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Trevor W. Stone
Wahyu Surya, Yan Li, Jaume Torres
School of Biological Sciences, Nanyang Technological University, Singapore
Abstract: A family of small polypeptides in many virus types associate to form oligomers and have channel activity. These proteins have been referred to as viroporins or virochannels and are increasingly recognized as important virulence factors and potential drug targets. In this review, we focus on two of the viroporins that have been studied in more detail from a structural and functional point of view. One is the 76-residue envelope (E) protein found in coronaviruses (CoVs) that causes the severe acute respiratory syndrome (SARS). The other is the 65-residue small hydrophobic (SH) protein found in a paramyxovirus, the respiratory syncytial virus (RSV). RSV SH and SARS-CoV E proteins are short polypeptides with a single transmembrane domain. In both cases, the presence of the viroporin has a protective effect on cells, preventing early apoptosis, but it leads to increased virulence in infected animal models. Both viroporins form homopentameric oligomers that show channel activity with no or low selectivity. The role of channel activity is still unclear, but associations have been made to facilitation of the egress of the virus by modification of the secretory pathway, and contributions to inflammation. SARS-CoV E protein has a cytoplasmically oriented C-terminus and a lumenal N-terminus, whereas the opposite orientation is found in RSV SH protein. Despite this opposite topology, nuclear magnetic resonance (NMR)-based structural models of these two channels show a similar champagne flute shape, with the wider opening facing the cytoplasmic side. Good channel inhibitors are lacking, but those found seem to have a preference for the narrow end of the channel. Availability of good inhibitors will help reveal the specific role of these channels in the life cycle of these viruses.
Keywords: respiratory syncytial virus, severe acute respiratory syndrome, small hydrophobic protein, envelope protein, channel structure
Viroporins
The term “viroporins” was proposed about 20 years ago after the observation of enhanced membrane permeability in virus-infected cells.1 Viroporins2,3 are a class of small proteins (~60–120 amino acids long) with one or two hydrophobic transmembrane (TM) domains that oligomerize to produce ion channels (hydrophilic pores) at the membranes of the host cell. In addition to the best studied case, the 97-residue M2 protein in influenza A,4–6 many viruses encode viroporins, for example, p7 in the hepatitis C virus (HCV),7 Vpu in human immunodeficiency virus type 1,8 2B in picornavirus,9 6K in alphavirus,10 p10 in avian reovirus,11 p7 in bovine viral diarrhea virus,12 Kcv in paramecium bursaria chlorella virus 1,13 and the list of members of this class of viral proteins keeps growing.7–20 The two that will be the focus of our review, the small hydrophobic (SH) protein in the respiratory syncytial virus (RSV),18 and envelope (E) proteins in coronaviruses (CoVs),21,22 are among the very few that have been studied structurally in some detail.
Envelope proteins in CoVs
CoVs are enveloped viruses organized into three groups:23 group 1 (α-CoVs) includes the porcine transmissible gastroenteritis virus (TGEV) and human CoV 229E, group 2 (β-CoVs) includes the murine hepatitis virus (MHV), severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV, and group 3 (γ-CoV) includes the avian infectious bronchitis virus (IBV). Representative sequences of these groups are shown in Figure 1A. Of particular medical interest are the SARS-CoV, which produced a near pandemic in 2003, and the recently emerged MERS-CoV, with 837 confirmed cases and at least 291 deaths (http://www.who.int/csr/don/2014_07_23_mers/en/).
CoV E proteins are short (76–109 amino acids) structural membrane proteins present at low concentration in the CoV virion,20,24–26 but found abundantly in internal membranes of infected cells, from ER to Golgi.20,27–29 For example, MHV E and SARS-CoV E are found localized at the ER-Golgi intermediate compartment, where CoV virions are assembled and bud into the lumen.30,31
E proteins have a predicted α-helical TM domain (Figure 1B), with two or three cysteine residues at the juxtamembrane region. Determination of the topology of E proteins has been confused by the fact that epitope tags can affect the topology of these small proteins.32–34 However, a recent study31 of untagged SARS-CoV E protein showed NexoCcyto topology in infected cells, that is, a cytoplasmic C-terminal domain and a lumenal N-terminus, and this topology is also likely in other E proteins.26,29,31,32
Deletion of E protein (ΔE) is deleterious or otherwise attenuating to the virus.35,36 SARS-CoV ΔE, for example, grows to a lower titer and shows aberrantly shaped virions.37 In vivo, this virus is highly attenuated,38 which has led to the development of a live vaccine.39 This route seems to be highly promising based on mice trial results40 and has also been suggested as a way to curb the spread of MERS-CoV.41
Infected cells by SARS-CoV ΔE underwent apoptosis more rapidly and to greater extent, showed upregulation of stress response genes, downregulation of inflammation genes,42 and decreased nuclear factor kappa-light-chain-enhancer of activated B cells-mediated inflammation response in infected cells and mice.43
Structure of the envelope protein in SARS-CoV
The SARS-CoV E protein structure was initially investigated using synthetic peptides corresponding to its predicted TM domain and molecular dynamics simulations.44 A pentameric model was proposed that was later confirmed using infrared linear dichroism data from 13C=18O isotopically labeled synthetic peptides incorporated in lipid bilayers.45,46 While cross-linking studies with full-length tagged SARS-CoV E-protein indicated undefined oligomers,20,21 the pentameric nature of the oligomer has been confirmed using analytical ultracentrifugation of the full-length polypeptide.47,48 Current data suggest that disulfide bonds, or the presence of juxtamembrane cysteines, are not required for oligomerization for TGEV E,24 IBV E,47 MHV E,30 or SARS E.48
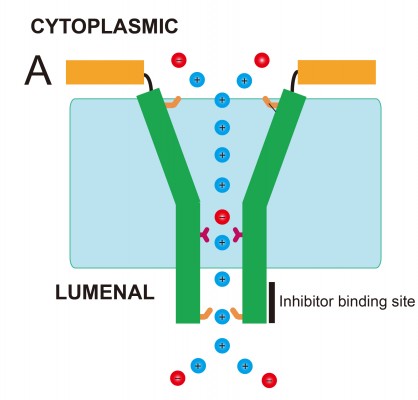
A more detailed study of the TM structure of SARS-CoV E was obtained from solution NMR of a synthetic isotopically labeled E (8–38) in dodecylphosphocholine (DPC) detergent micelles.19 A model of the pentamer was derived from intermonomeric Nuclear Overhauser effects and paramagnetic relaxation enhancement data, confirming the TM helix orientations reported previously.45,46 Although the resolution of the details is still low, these data produced a consistent picture of the channel geometry, where Val25 is located at the narrowest point of the lumen (Figure 2A).
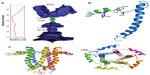  | Figure 2 NMR-based models of SARS-CoV E proteins. |
Interestingly, the predicted β-structure region, or hairpin, centered on a totally conserved Pro residue (Figure 1), formed 100% β structure when studied as a short peptide,49 but was found to be mostly α-helical in the context of a longer construct E (8–65; Figure 2B) and also full-length SARS-CoV E.48 This α-helical conformation is consistent with circular dichroism and Fourier transform infrared spectroscopy data.47 Overall, this discrepancy may point to the existence of a frustrated structure poised for conformational change in this part of the molecule. These data have resulted in a mostly α-helical model for SARS-CoV E that is reminiscent of that of HCV p7 (Figure 2C).50
Channel activity of SARS-CoV E and its pathological role
Channel activity with very mild cation selectivity has been observed in black lipid membrane (BLM) systems using E protein synthetic peptides encompassing their predicted TM region.51,52 Later studies have shown minor changes in selectivity induced by lipid charge53 and pH,54 and the N-terminal Glu residues in SARS-CoV E have been suggested to confer some selectivity.54 A truncated form of SARS-CoV E (8–65) in DPhPC (diphytanoylphosphatidylcholine) showed a conductance of 0.39±0.02 nS.48 For comparison, synthetic full-length SARS-CoV E and E (7–38) produced single channel conductances of 0.19±0.06 nS and 0.18±0.12 nS in 1 M NaCl,53 respectively. The lower conductance observed in synthetic samples may be due to extraneous modifications or impurities resulting from exposure to harsh chemicals.
Despite initial patch clamp data reports showing cation selective channel activity,19 SARS-CoV E protein could not be detected at the plasma membrane of transfected cells31 or in frog oocytes.55 In the latter system, co-expression of SARS-CoV E with human epithelial sodium transporter in Xenopus oocyte decreased amiloride-sensitive current through activation of protein kinase C and subsequent decrease in epithelial sodium transporter surface levels.55 A similar direct or indirect inhibitory effect on other endogenous channels was observed by patch clamp in transfected cells.31
Early indications showed that channel activity of E proteins may be important for CoVs. For example, titer, plaque size, and hexamethylene amiloride (HMA) sensitivity was reduced when parts of the TM were rotated by insertion of Ala residues in various positions.56 Synthetic peptides of SARS-CoV E carrying several mutations have contributed to identify residues that abolish ion channel activity, that is, Val25 and Asn15.57 The latter is in a conserved position in the TM domain of all E proteins, were a polar residue is always found (Figure 1A). The side chains of these two residues face the lumenal side and monomer–monomer interface, respectively.58
Consistently, SARS-CoV carrying E protein with channel-inactivating N15A and V25F mutations was attenuated in vivo and led to the appearance of compensatory mutations that rapidly outcompeted the inactive mutants.59 Mutation N15A was compensated by A15D, whereas pentamer-disruptive mutation V25F was compensated at the neighbor α-helix in the pentameric channel that faces residue 25.19,63 This is consistent with the primary destabilization of the pentameric oligomer by V25F. Mutation N15A, in contrast, does not destabilize the oligomer (results unpublished); therefore, inhibition may be related to a more fundamental aspect of channel activity and interaction with the ions.
These revertant mutants restored ion channel activity and mortality comparable to wild-type (WT), showing for the first time that ion channel activity of E protein is a determinant of CoV pathogenesis.59 The precise mechanism by which SARS-CoV E protein channel activity contributes to virulence is still not clear, but it may be related to inflammasome activation and elevation of interleukin-1β.59
Also, during virion egress, CoV infection drives a rearrangement of host cell membranes including the Golgi complex,60 and virions appear in large vacuoles derived from Golgi/ER-Golgi intermediate compartment membranes.61 When IBV E residue Thr16 was mutated to Ala, this resulted in decreased Golgi disruption.62 Thr16 is aligned with SARS-CoV E Asn15 (Figure 1) and therefore is expected to abolish channel activity. Thus, in addition to its possible contribution to virion assembly, channel activity of E proteins may contribute to CoV virulence by disrupting the balance of lumenal environments and subsequently affecting the secretory pathway, leading to release of virions.63 However, this disruptive effect could not be restored by conservative polar substitutions with Ser, Asn, or Gln;32 therefore, other context-dependent changes may be necessary to recover channel activity.
Human RSV SH protein
The human respiratory syncytial virus (hRSV) is an enveloped pneumovirus in the paramyxoviridae family that causes lower respiratory tract disease in infants, elderly, and immunocompromised populations worldwide.64 Up to 64 million reported cases of hRSV infection and 160,000 deaths occur each year. Although the virus was identified almost half a century ago,65 there are still no vaccines or effective antiviral drugs available.
The SH protein is one of the three RSV membrane proteins, along with F and G, which are key factors during virus attachment, fusion, and entry into host cells.66,67 The SH protein is a type II integral membrane protein 64 (RSV subgroup A) or 65 (RSV subgroup B) amino acids long, with a single α-helical TM domain68,69 (Figure 3A) which is highly conserved.70,71 The C- and N-terminal extramembrane domains are oriented lumenally/extracellularly and cytoplasmically, respectively, that is, opposite to SARS-CoV E. Most SH protein accumulates at the membranes of the Golgi complex in infected cells, but it has also been detected in the endoplasmic reticulum and plasma membranes.72 During infection, the full-length unmodified form is the major species.68
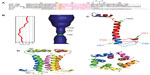  | Figure 3 Sequences of hRSV SH protein and homologs in paramyxoviruses. |
RSV lacking the SH gene (RSVΔSH) is viable, causes formation of syncytia, and grows as well as the WT virus in cell culture.73–76 However, RSVΔSH virus is attenuated and replicates tenfold less efficiently than the WT in the upper respiratory tract.75,76 RSVΔSH virus is also attenuated in vivo by virus passage in mouse and chimpanzee models.73,77 Overall, these results indicate involvement of SH protein in the pathogenesis of RSV infection.
In common to SARS-CoV E protein, SH protein blocks or delays apoptosis in infected cells. This protective effect is also observed in homologs of RSV SH protein (Figure 3A), that is, parainfluenza virus 5, mumps virus, and J paramyxovirus.74,78–80 Recent reports have also suggested that SH protein activates the NLRP3 inflammasome.81
Structure of RSV SH protein
Earlier cross-linking studies showed that SH protein form different oligomers in sodium dodecyl sulphate, including dimers, trimers, tetramers, and pentamers.68,82 However, both synthetic TM domain (residues 18–43) and full-length SH protein have been shown to form homopentamers in a variety of detergents.75,83 These oligomers may have been responsible for early reports showing increased entry in bacteria of low molecular weight compounds after SH protein expression.18
The mutual orientation of monomers in the TM pentamer was determined using site-specific infrared dichroism of isotopically labeled peptides in lipid bilayers.83 hRSV SH protein has two Histidine (His) residues, His22 and His51 (Figure 3A). These structural studies showed His22 to be in a lumenal, close to interhelical, orientation. This orientation was later confirmed by NMR studies that reconstructed the pentameric α-helical bundle of the full-length protein in DPC micelles.75 Like in SARS-CoV E, the pore lumen is more open toward the cytoplasmic side (Figure 3B). In this “micelle” model, a single α-helical TM domain was flanked N-terminally by an α-helix, and C-terminally by an extended β-hairpin. Later, the structure of these extramembrane domains has been confirmed using a DHPC/DLPC (1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine/1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine) bicellar system.84 However, in that “bicelle” model, the TM α-helix was extended one more turn, until the other His residue, His51, whereas Pro-58 is located at the tip of a C-terminal loop (Figure 3C).
Using the mutual orientations between TM domains in lipidic membranes and DPC micelles,83,75 the pentameric model of the SH protein channel was reconstructed (Figure 3D). According to this model, His51 side chains are facing the lumen of the channel, whereas His22 is slightly skewed toward the other helix–helix interface.
Channel activity of RSV SH protein
The contribution to channel activity and selectivity of these two His residues was examined using BLMs and purified SH protein. Although the overall conductance was pH-dependent, consistent with previous patch clamp reports,75 SH protein showed lower conductance, not activation, at low pH. Indeed, conductance was consistent with the titration of His residues, with less conductance at low pH, where both His should be protonated. Also, His residues are not required for channel activity, as the channel was active even when both His residues were changed to Phe.84 These results are consistent with a dramatic reduction of channel activity at lower pH observed previously for synthetic SH-TM (residues 18–43) in planar lipid bilayers,83 where only His22 was present. Overall, BLM results showed only minor variations in conductance and selectivity with pH.84 Similarly, ion selectivity was low, and appeared modulated by His51, with His22 contributing a more structural role. This is consistent with the observed electrophoretic pattern of His mutants;75 mutant H22A showed aberrant mobility and/or aggregation, whereas H51A showed mobility similar to the WT form. Poor in vitro ion selectivity has also been observed in other viroporins, for example, SARS-CoV E protein,53 or HCV p7,85 and it may be a hallmark of incomplete specialization of these channels.
Channel activity inhibitors of SARS-CoV E and RSV SH proteins
Availability of channel inhibitors would help in understanding the role of membrane permeabilization to ions in the viral life cycle of these viruses, and would also have obvious therapeutic potential. SARS-CoV E channel activity is inhibited partially by HMA in the Kb~10 μM range.51,52 The interaction of this drug with the channel has been mapped to residues located at the N- and C-terminal ends of the TM region48,19 (Figure 4A and B). Affected residues at the N-terminal end include the conserved polar residue Asn-15, suggesting HMA binding to the N-terminal, luminal facing, end of the channel.
  | Figure 4 Binding sites of inhibitors in SARS-CoV E and SH proteins. (A) Superposition of TROSY-HSQC spectra of uniformly 15N-labeled SARS-CoV E in the absence (red) and presence (blue) of 0.4 mM HMA (shown above image A). Peaks that undergo significant shifts upon complex formation are highlighted; (B) residues showing larger and smaller shifts are represented in red and yellow, respectively; (C) same as (A) for uniformly 15N-labeled SH protein in the absence (red) and presence (blue) of 4.8 mM pyronin B (shown above image C). (D) Residues showing larger and smaller shifts are represented in red and yellow, respectively. |
RSV SH protein is partially inhibited by pyronin B with a binding affinity (Kd) of ~7 μM.84 Although modest, this Kd compares favorably with the 16 μM for amantadine inhibition of influenza M2 protein,86 the ~10 μM reported for HMA inhibition of SARS-CoV E,52 50–100 μM for rimantadine inhibition of HCV p7,50 or the >100 μM of HMA for p7 inhibition.85 SH protein conductance changed from 0.3±0.07 nS in control conditions to 0.12±0.06 nS in the presence of pyronin B. As in the case of HMA for SARS-CoV E (Figure 4A and B), binding of pyronin B was mapped at both ends of the TM domain (Figure 4C and D). At the C-terminal end, the residue most affected was Ala39, and a group of nearby residues. Interestingly, most of these C-terminal juxtamembrane residues (residues 38–43) form a conserved motif in RSV SH protein “A39ILNKL43,” suggesting that binding of the drug may not only alter channel activity. This binding site is located at the luminal-facing narrowest region of the channel lumen (Figure 3B). By comparison, residues affected at the N-terminal end are not conserved. Pyronin B was found to bind SH protein mostly at the lipid-facing side of the TM α-helices and not into the pore lumen. This mechanism of inhibition is thus probably allosteric, similar to that proposed for rimantadine binding to the TM domain of M250 or rimantadine to HCV p7.50
Conclusion
In summary, despite the sequences of SARS-CoV E and hRSV SH protein not being evolutionarily related, they share remarkable structural and functional features. Both viroporins have a single α-helical TM domain that forms pentameric oligomers. These oligomers are responsible for channel activity that is not selective. In both cases, the TM α-helix is connected to a short cytoplasmic membrane-bound α-helix by a flexible loop (Figure 5). The lumen of the pore has a funnel-shaped structure, more open toward the cytoplasmic side. The luminal side is narrower and contains conserved residues that may be important for channel activity, that is, Asn-15 in SARS-CoV E and His51 in RSV SH protein. This narrower luminal end is also where inhibitors were found to bind.48,84 This funnel-like architecture has also been observed in other viroporins, for example, influenza M2 protein5 and HCV p7.50
Both viroporins delay apoptosis in infected cells, which may help evade host inflammatory responses and the premature death of the host cells. However, their presence increases pathogenicity. The relation of channel activity with virulence has been established at least for SARS-CoV E. Hence, viroporins constitute an as yet untapped potential source of pharmacological targets.
In addition to this channel activity, both E and SH proteins have been found to participate in a number of interactions with viral and host proteins that are not the subject of this article, for example, SARS-CoV E interacts with viral non-structural protein 3,87 M protein,29,88–90or host PALS1,88 whereas SH protein interacts with viral G protein.89,90 These interactions probably contribute to viral morphogenesis and viral particle formation and participating in the entry or release of viral particles into or from cells.
Acknowledgments
This work has been funded by the Singapore National Research Foundation under CRP Award No NRF-CRP4-2008-02 and Tier 1 grant RG 51/13.
Author contributions
All authors contributed to conception and design, acquisition of data, or analysis, and interpretation of data; drafting the article or revising it critically for intellectual content; and final approval of the version to be published.
Disclosure
The authors report no conflicts of interest in this work.
References
Carrasco L, Sanz MA, Ayala MJ, et al. Modification of membrane permeability by animal viruses. In: Carrasco L, Sonenberg N, Wimmer E, editors. Regulation of Gene Expression in Animal Viruses. Vol 40. New York: Plenum Press; 1993:283–305. | |
Gonzalez ME, Carrasco L. Viroporins. FEBS Lett. 2003;552(1):28–34. | |
Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol. 2012;10(8):563–574. | |
Stouffer AL, Acharya R, Salom D, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451(7178):596–599. | |
Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451(7178):591–595. | |
Pielak RM, Schnell JR, Chou JJ. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci U S A. 2009;106(18):7379–7384. | |
Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39(1):5–19. | |
Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398(1):12–18. | |
Cuconati A, Xiang W, Lahser F, Pfister T, Wimmer E. A protein linkage map of the P2 nonstructural proteins of poliovirus. J Virol. 1998;72(2):1297–1307. | |
Sanz MA, Pérez L, Carrasco L. Semliki forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J Biol Chem. 1994;269(16):12106–12110. | |
Bodelon G, Labrada L, Martinez-Costas J, Benavente J. Modification of late membrane permeability in avian reovirus-infected cells: viroporin activity of the S1-encoded nonstructural p10 protein. J Biol Chem. 2002;277(20):17789–17796. | |
Harada T, Tautz N, Thiel HJ. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J Virol. 2000;74(20):9498–9506. | |
Plugge B, Gazzarrini S, Nelson M, et al. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287(5458):1641–1644. | |
Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. | |
Melton JV, Ewart GD, Weir RC, Board PG, Lee E, Gage PW. Alphavirus 6K proteins form ion channels. J Biol Chem. 2002;277(49):46923–46931. | |
Chen MY, Maldarelli F, Karczewski MK, Willey RL, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: The cytoplasmic domain of CD4 contributes to Vpu sensitivity. J Virol. 1993;67(7):3877–3884. | |
Lamb RA, Holsinger LJ, Pinto LH. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Receptor-Mediated Virus Entry into Cells. Vol 5. Long Island, NY: Cold Spring Harbor Laboratory Press; 1994:303–321. | |
Perez M, Garcia-Barreno B, Melero JA, Carrasco L, Guinea R. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology. 1997;235(2):342–351. | |
Pervushin K, Tan E, Parthasarathy K, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Path. 2009;5(7):e1000511. | |
Liao Y, Yuan Q, Torres J, Tam JP, Liu DX. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349(2):264–275. | |
Liao Y, Lescar J, Tam JP, Liu DX. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem Biophys Res Commun. 2004;325(1):374–380. | |
Madan V, Garcia Mde J, Sanz MA, Carrasco L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 2005;579(17):3607–3612. | |
Enjuanes L, Brian D, Cavanagh D, et al. Coronaviridae. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, et al, editors. Virus Taxonomy. Classification and Nomenclature of Viruses. San Diego: Academic Press; 2000:835–849. | |
Godet M, L’Haridon R, Vautherot J-F, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188(2):666–675. | |
Yu X, Bi W, Weiss SR, Leibowitz JL. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202(2):1018–1023. | |
Corse E, Machamer CE. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol. 2000;74(9):4319–4326. | |
Tung FYT, Abraham S, Sethna M, et al. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186(2):676–683. | |
Corse E, Machamer CE. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312(1):25–34. | |
Raamsman MJ, Locker JK, de Hooge A, et al. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74(5):2333–2342. | |
Lopez LA, Riffle AJ, Pike SL, Gardner D, Hogue BG. Importance of conserved cysteine residues in the coronavirus envelope protein. J Virol. 2008;82(6):3000–3010. | |
Nieto-Torres JL, Dediego ML, Alvarez E, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. | |
Ruch TR, Machamer CE. A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein. PLoS Pathog. 2012;8(5):e1002674. | |
Maeda J, Repass JF, Maeda A, Makino S. Membrane topology of coronavirus E protein. Virology. 2001;281(2):163–169. | |
Yuan Q, Liao Y, Torres J, Tam JP, Liu DX. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006;580(13):3192–3200. | |
Ortego J, Escors D, Laude H, Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J Virol. 2002;76(22):11518–11529. | |
Ortego J, Ceriani JE, Patino C, Plana J, Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368(2):296–308. | |
DeDiego ML, Alvarez E, Almazán F, et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81(4):1701–1713. | |
DeDiego ML, Pewe L, Alvarez E, Rejas MT, Perlman S, Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376(2):379–389. | |
Lamirande EW, DeDiego ML, Roberts A, et al. A live attenuated severe acute respiratory syndrome coronavirus is immunogenic and efficacious in golden Syrian hamsters. J Virol. 2008;82(15):7721–7724. | |
Netland J, DeDiego ML, Zhao J, et al. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399(1):120–128. | |
Almazán F, Dediego ML, Sola I, et al. Engineering a replication-competent, propagation-defective middle east respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4(5):e613–e650. | |
DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, et al. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Path. 2011;7(10):e1002315. | |
DeDiego ML, Nieto-Torres JL, Regla-Nava JA, et al. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. 2014;88(2):913–924. | |
Torres J, Wang J, Parthasarathy K, Liu DX. The transmembrane oligomers of coronavirus protein E. Biophys J. 2005;88(2):1283–1290. | |
Torres J, Parthasarathy K, Lin X, Saravanan R, Kukol A, Ding XL. Model of a putative pore: the pentameric α-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys J. 2006;91(3):938–947. | |
Parthasarathy K, Ng L, Lin X, et al. Structural flexibility of the pentameric SARS coronavirus envelope protein ion channel. Biophys J. 2008;95(6):L39–L41. | |
Parthasarathy K, Lu H, Surya W, Vararattanavech A, Pervushin K, Torres A. Expression and purification of coronavirus envelope proteins using a modified beta-barrel construct. Prot Exp Purif. 2012;85(1):133–141. | |
Li Y, Surya W, Claudine S, Torres J. Structure of a conserved golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem. 2014;289(18):12535–12549. | |
Surya W, Samsó M, Torres J. Structural and functional aspects of viroporins in human respiratory viruses: respiratory syncytial virus and coronaviruses. In: Vats M, editor. Respiratory Disease and Infection – A New Insight. Croatia: InTech; 2013:47–76. | |
OuYang B, Xie S, Berardi MJ, et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498(7455):521–525. | |
Wilson L, McKinlay C, Gage P, Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330(1):322–331. | |
Wilson L, Gage P, Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. | |
Verdiá-Báguena C, Nieto-Torres JL, Alcaraz A, et al. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. | |
Verdiá-Báguena C, Nieto-Torres JL, Alcaraz A, Dediego ML, Enjuanes L, Aguilella VM. Analysis of SARS-CoV e protein ion channel activity by tuning the protein and lipid charge. Biochim Biophys Acta. 2013; 1828(9):2026–2031. | |
Ji HL, Song W, Gao Z, et al. SARS-CoV proteins decrease levels and activity of human ENaC via activation of distinct PKC isoforms. Am J Physiol – Lung Cell Mol Physiol. 2009;296(3):L372–L383. | |
Ye Y, Hogue BG. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J Virol. 2007;81(7):3597–3607. | |
Torres J, Maheswari U, Parthasarathy K, Ng L, Ding XL, Gong X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16(9):2065–2071. | |
Torres J, Parthasarathy K, Lin X, Saravanan R, Kukol A, Liu DX. Model of a putative pore: the pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys J. 2006;91(3):938–947. | |
Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014; 10(5):e1004077. | |
Lavi E, Wang Q, Weiss SR, Gonatas NK. Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology. 1996;221(2):325–334. | |
Ulasli M, Verheije MH, de Haan CAM, Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12(6):844–861. | |
Ruch TR, Machamer CE. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J Virol. 2011;85(2):675–685. | |
Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4(3):363–382. | |
Dowell SF, Anderson LJ, Gary HE Jr, et al. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J Infect Dis. 1996;174(3):456–462. | |
Blount RE Jr, Morris JA, Savage RE. Recovery of cytopathogenic agent from chimpanzees with coryza. Proc Soc Exp Biol Med. 1956;92(3):544–549. | |
Krusat T, Streckert HJ. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142(6):1247–1254. | |
Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197(1):1–11. | |
Collins PL, Mottet G. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J Gen Virol. 1993;74:1445–1450. | |
Gan SW, Tan E, Lin X, et al. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem. 2012;287(29):24671–24689. | |
Collins PL, Olmsted RA, Johnson PR. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J Gen Virol. 1990;71:1571–1576. | |
Chen MD, Vazquez M, Buonocore L, Kahn JS. Conservation of the respiratory syncytial virus SH gene. J Infect Dis. 2000;182(4):1228–1233. | |
Rixon HW, Brown G, Aitken J, McDonald T, Graham S, Sugrue RJ. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J Gen Virol. 2004;85(pt 5):1153–1165. | |
Bukreyev A, Whitehead SS, Murphy BR, Collins PL. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71(12):8973–8982. | |
Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the respiratory syncytial virus small hydrophobic protein. J Virol. 2007;81(15):8361–8366. | |
Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology. 2000;273(1):210–218. | |
Karron RA, Buonagurio DA, Georgiu AF, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Nat Acad Sci U S A. 1997;94(25):13961–13966. | |
Whitehead SS, Bukreyev A, Teng MN, et al. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73(4):3438–3442. | |
Lin Y, Bright AC, Rothermel TA, He B. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J Virol. 2003;77(6):3371–3383. | |
Wilson RL, Fuentes SM, Wang P, et al. Function of small hydrophobic proteins of paramyxovirus. J Virol. 2006;80(4):1700–1709. | |
Li Z, Xu J, Patel J, et al. Function of the small hydrophobic protein of J paramyxovirus. J Virol. 2011;85(1):32–42. | |
Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax. 2013;68(1):66–75. | |
Rixon HWM, Brown G, Murray JT, Sugrue RJ. The respiratory syncytial virus small hydrophobic protein is phosphorylated via a mitogen-activated protein kinase p38-dependent tyrosine kinase activity during virus infection. J Gen Virol. 2005;86(2):375–384. | |
Gan SW, Ng L, Lin X, Gong X, Torres J. Structure and ion channel activity of the human respiratory syncytial virus (hRSV) small hydrophobic protein transmembrane domain. Protein Sci. 2008;17(5):813–820. | |
Li Y, To J, Verdià-Baguena C, et al. Inhibition of the human respiratory syncytial virus small hydrophobic protein and Structural variations in a bicelle environment. J Virol. 2014;88(20):11899–11914. | |
Premkumar A, Wilson L, Ewart GD, Gage PW. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557(1–3):99–103. | |
Jing X, Ma C, Ohigashi Y, et al. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Nat Acad Sci U S A. 2008;105(31):10967–10972. | |
Alvarez E, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Marcos-Villar L, Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402(2):281–291. | |
Teoh KT, Siu YL, Chan WL, et al. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21(22):3838–3852. | |
Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001;75(15):6825–6834. | |
Low KW, Tan T, Ng K, Tan BH, Sugrue RJ. The RSV F and G glycoproteins interact to form a complex on the surface of infected cells. Biochem Biophys Res Commun. 2008;366(2):308–313. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.