Back to Journals » Infection and Drug Resistance » Volume 10
Pegylated-interferon plus ribavirin treatment does not alter the prevalence of resistance-associated substitutions to direct-acting antivirals in HCV genotype 1a patients
Authors Chen Z, Pang X, Li Z, Ren H , Hu P
Received 2 July 2017
Accepted for publication 10 August 2017
Published 31 August 2017 Volume 2017:10 Pages 275—281
DOI https://doi.org/10.2147/IDR.S145362
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Zhi-wei Chen,* Xi-chen Pang,* Zhao Li, Hong Ren, Peng Hu
Department of Infectious Diseases, Institute for Viral Hepatitis, The Key Laboratory of Molecular Biology for Infectious Diseases, Chinese Ministry of Education, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China
*These authors contributed equally to this work
Background: Direct-acting antiviral (DAA) resistance-associated substitutions (RASs) can jeopardize the effectiveness of DAAs in patients with hepatitis C virus (HCV). The selection pressure by pegylated-interferon (Peg-IFN) plus ribavirin (P/R) treatment may enhance HCV genome variation. However, whether P/R treatment alters the rate of change of RASs is still unclear.
Materials and methods: We retrieved the genomic sequences of HCV genotype (GT) 1a patients from GenBank, which included patients naïve to P/R (pre-IFN group) and those previously treated with P/R (post-IFN group). The sequences were aligned and analyzed by using MEGA 6.0 software. Clinically relevant RASs were summarized from the current medical literature.
Results: In the cross-sectional study, the total prevalence of clinically relevant RASs was high, independent of the treatment group (pre-IFN: 219/403 [54.34%] vs post-IFN: 67/131 [51.15%]). The high prevalence was mainly detected in the NS3 region RAS at Q80 (40.69% vs 36.64%). The RASs in the NS5A region, such as M28, Q30, L31 and Y93, were uncommon (0%–5%). Similarly, all RASs showed no difference between the two groups. One exception was the RAS at I170 in the NS3 region, which was significantly higher in the post-IFN group than in the pre-IFN group. In the longitudinal study, similar results were observed. However, no difference in RAS at I170 was observed between the two groups. Finally, no clinically relevant RASs were detected in response to the DAA regimens approved for GT 1a patients treated with P/R.
Conclusion: Our results suggest that previous P/R treatment failure was not favorably associated with an increase in DAAs RASs present in GT1a patients. Our results support the American Association for the Study of Liver Diseases’ recommendations of DAA intervention in P/R-treated GT1a patients.
Keywords: pegylated-interferon, ribavirin, direct-acting antivirals, resistance-associated substitutions, prevalence
Introduction
Infection with the hepatitis C virus (HCV) is a serious global public health problem, with more than 170 million individuals infected worldwide.1 The combination of pegylated-interferon (Peg-IFN) plus ribavirin (P/R), as a standard of care for HCV-infected patients, induces a sustained virologic response (SVR) in only 40%–50% of patients with genotype (GT) 1 infection.2,3 To achieve a higher SVR rate, better tolerability and shorter treatment duration, direct-acting antiviral agents (DAAs) have been developed recently, which specifically target three key regions of the HCV genome.4 Since the first DAA was approved in 2011, it has opened a new era in HCV treatment. For most GT1 HCV patients who did not achieve SVR with P/R treatment, DAAs are a better choice.
However, due to the low fidelity of polymerase, high replication rate and selection pressures on HCV, HCV exists as a collection of viruses named “quasispecies”.5 HCV variants that reduce the susceptibility to DAAs have been called resistance-associated substitutions (RASs). RASs are a major limitation in the development of new DAAs. DAA-resistant phenotypes have been observed both in vitro and during clinical trials.6,7 As some RASs have been associated with DAA treatment failure, we defined them as clinically relevant RASs.
Peg-IFN can enhance the host-specific antiviral immune response and clear the infected cells.8 As a result of elevated immunologic pressure by Peg-IFN, HCV variants may become more diversified to avoid the host immune response. In addition, a previous study suggested that ribavirin is an RNA mutagen, causing accumulation of HCV mutations during replication.9 Consequently, we hypothesized that some RASs could also occur as a hitchhiking effect during P/R treatment. However, the nature of the RASs, especially of clinically relevant RASs, in P/R treatment failure in GT1 patients remains unclear.
In this cross-sectional and longitudinal study, we analyzed the frequency of clinically relevant RASs in P/R treatment failure in HCV GT1a patients based on the HCV genomic sequences from GenBank.
Materials and methods
Patient sequence screening strategy
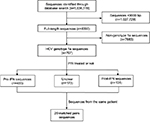
As described previously, HCV genomic sequences were retrieved from GenBank (www.ncbi.nlm.nih.gov/nuccore) in June 2017 using the key words “hepatitis C virus” or “HCV”. After an initial search, near full-length HCV sequences (>9000 bp) were screened. Each sequence GT was retrieved and identified with the NCBI viral genotyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi) and non-GT1a HCV sequences were excluded. Finally, detailed information regarding the P/R treatment was obtained based on the annotation of each HCV GT1a sequence from GenBank (Figure 1).
Variant analyses and definition
Clinically relevant RASs of all approved DAAs included in this study were identified from the most recently available publications and are summarized in Table 1.10–21 Sequences were aligned and analyzed with MEGA 5.0 software (Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA). Variant type was described as a substitution of the consensus amino acid with a novel one compared with the reference sequence (accession number: NC. 004102). For instance, Y93H and Y93N in the NS5A region were described as two variant types.
Statistical analyses
All data are presented as rates (%) and were analyzed statistically using chi-square test with SPSS 17 software (SPSS Inc., Chicago, IL, USA). P values were calculated with two-tailed statistical analysis, and P values <0.05 were considered statistically significant.
Results
Screening of HCV GT1a sequences
We identified 1,036,119 sequences in the NCBI Nucleotide Database as of June 2017 using the key words “hepatitis C virus” or “HCV”. After removing the HCV sequences with <9000 bp, we narrowed the list of sequences of interest to 8390 sequences. Further excluding HCV non-GT1a sequences by use of online genotyping tools as described in the “Materials and methods” section, we obtained 707 HCV GT1a sequences. Detailed P/R treatment information for each selected sequence was retrieved according to its annotation in GenBank. We identified 403 sequences that were derived from HCV patients who were naïve to P/R treatment (defined as the pre-IFN group) and 131 sequences that were derived from HCV patients after P/R treatment failure (defined as the post-IFN group). In the pre-IFN group and post-IFN group, we still found 20 pairs of HCV sequences, and each of them was derived from the same patient (Figure 1). GenBank accession numbers of HCV sequences used in this study are provided in Table S1.
Prevalence of clinically relevant RASs in a cross-sectional study
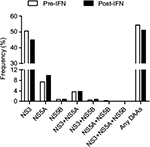
To determine the prevalence of clinically relevant RASs in P/R treatment failure patients and whether P/R treatment has an effect on these RASs in a cross-sectional study, we compared the frequency of RASs in the pre-IFN group and post-IFN group. First, the overall prevalence of clinically relevant RASs in the post-IFN group was high (67/131, 54.31%). However, the difference between the pre-IFN and post-IFN groups was not statistically significant (Figure 2). The high prevalence of clinically relevant RASs was mainly detected in the NS3 region, followed by the NS5A region and NS3+NS5A region (45.04%, 9.92% and 3.82%, respectively). Clinically relevant RASs were rarely observed in the NS5B region and NS3+NS5B region (<1%). Remarkably, no clinically relevant RASs were found in the NS5A+NS5B region and all three combined regions (Figure 2). Similarly, no significant difference was observed in these regions between the two groups.
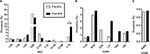
We also analyzed the change in specific clinically relevant RASs in each HCV region. In the NS3 region, the highest frequency of clinically relevant RASs was at Q80 (resistant to simprevir), but the difference between the two groups was not significant (40.69% vs 36.64%; Figure 3A). Similarly, the difference in RASs at V36, T54, V55, S122, I132, R155 and D168 was not significant. However, the prevalence of RASs at I170 (resistance to boceprevir) was significantly higher in the post-IFN group than that in the pre-IFN group (3.23% vs 9.92%, P<0.01; Figure 3A). In the NS5A region, the prevalence of key clinically relevant RASs at L31 (resistance to daclatasvir, ledipasvir and velpatasvir) and Y93 (resistance to daclatasvir, ledipasvir, velpatasvir and elbasvir) was low and no significant difference was observed between the two groups. Similar results were observed in the RASs at K24, M28, Q30 and H58 (Figure 3B). In the NS5B region, only one clinically relevant RAS was observed at S556 (resistance to dasabuvir). Its frequency was very low, and no significant difference was observed between the two groups as before (0.74% vs 0.75%, P>0.05; Figure 3C).
Finally, we investigated the prevalence of clinically relevant RASs to the approved DAA combination regimens: elbasvir + grazoprevir, ledipasvir + sofosbuvir, simeprevir + sofosbuvir, velpatasvir + sofosbuvir, daclatasvir + sofosbuvir and paritaprevir + ombitasvir + dasabuvir. As expected, no combination RAS was observed even in the P/R treatment failure patients (data not shown).
Prevalence of clinically relevant RASs in a longitudinal study
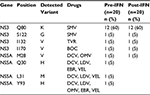
In the longitudinal study, clinically relevant RASs in 20 pairs of pre-IFN and post-IFN sequences were analyzed. Similar to the cross-sectional study, Q80K in the NS3 region was the clinically relevant RAS that was observed most commonly and the frequency was the same between the two groups (60% vs 60%). The remaining RASs, such as S122G and I132V in the NS3 region and M28V, Q30H, L31M and Y93H in the NS5A region, were rarely observed and no significant difference was observed between the two groups. No clinically relevant RAS was detected in the NS5B region in the 20 pairs of sequences. However, one RAS in the NS3 region was different from the cross-sectional study, that is, I170V. There was no significant difference between the pre-IFN and post-IFN groups (Table 2).
Discussion
Resistance to DAAs still hinders the cure of patients with HCV infection. Clinically relevant RASs play a main role in DAA resistance. However, little information is currently available about the changes in clinically relevant RASs in patients in whom P/R treatment failed. In this study, we found that even though a high prevalence of RASs was observed in P/R treatment failure patients, no changes occurred in the pre-IFN and post-IFN groups. No clinically relevant RASs were detected with DAA regimens that have been approved recently.
In the cross-sectional study, the overall prevalence of clinically relevant RASs was high, independent of the treatment group. This finding was consistent with our previous study.22 Regarding the treatment-naïve patients and those retreated after P/R treatment failure, the European Association for the Study of the Liver23 and the American Association for the Study of Liver Diseases (AASLD)24 both recommend baseline resistance testing for Q80K in the HCV GT1a genome when simeprevir is used in the treatment regimen. In this study, a high prevalence was mainly detected in RAS Q80K (40.69% vs 36.64%). The regional differences in the prevalence of RASs may account for the high frequency because all of the HCV sequences in our study were derived from patients in the USA. This finding was similar to that of a previous study,25 which also reported a high prevalence of Q80K in the USA (48%). Additionally, two patients of the pre-IFN group had an RAS observed at Q80R. It is worth noting that the prevalence of I107V (resistance to boceprevir) in the post-IFN group was significantly higher than in the pre-IFN group. This finding suggests that boceprevir should not be recommended to the P/R treatment failure patients. Furthermore, variants that confer low-level (V36M and R155K) resistance to telaprevir were observed with a low prevalence in our study, which is consistent with other findings.26–28 However, high-level (A156V/T, 36+155, 36+156) resistance to teleprevir26 was not detected in this study.
In patients with HCV GT1a infection, the presence of baseline NS5A RASs (such as M28, Q30, L31 and Y93) that cause a large reduction in the activity of NS5A inhibitors (>5-fold) adversely impacts the response to NS5A-containing regimens.24 In this study, these RASs were detected with a lower frequency (0%–4.58%). The prevalence of RASs at M28, L31 and Y93 seems to be higher in the post-IFN group than in the pre-IFN group, but the difference did not reach statistical significance. In the NS5B region, the key RAS S282T was not found in any patient, which is consistent with a previous study.29 In addition, only one RAS was observed at S556. Similarly, no significant difference was observed between the two groups.
In the longitudinal study, analogous with the cross-sectional study, no significant difference was observed between the pre-IFN group and post-IFN group. The results were consistent with a previous study,30 which found that P/R treatment failure does not alter the frequency of HCV NS3 and NS5B RASs in HCV/human immunodeficiency virus coinfected patients. However, the prevalence of RAS Q80K was higher in this study than in the earlier study (60% vs 27.78%).30 The different study patient populations may account for this difference.
Recommendation guidelines for DAA regimens in HCV GT1a patients treated with P/R have been published by the AASLD.24 As expected, no clinically relevant RASs were detected against these recommended DAA regimens. Therefore, our results support the AASLD recommendations of DAA regimens for HCV GT1a patients previously treated with P/R. In addition, several recent clinical studies have reported generally similar SVR rates in P/R treatment-naïve and P/R treatment-experienced patients across the various regimens.31–33 Our result that no difference in RASs was detected in the pre-IFN group and post-IFN group also provides support for the clinical findings.
This study offers two important strengths. First, we used HCV sequences from the current GenBank database to analyze the effect of P/R treatment on clinically relevant RASs both in cross-sectional and longitudinal studies. The number of HCV sequences in the cross-sectional study was large. Second, we analyzed full-length HCV sequences, such that we could investigate the combined effect of clinically relevant RASs in different regions at the same time, which was important in determining the multiresistance to DAA regimens recommended in the guidelines. However, this study presents some limitations as well. First, all HCV sequences were obtained from GenBank. Some information about patients was not integrated, which prevented further analyses. Second, the number of HCV sequences in the longitudinal study was small. Nevertheless, summarized from the cross-sectional and longitudinal study, the results were still reliable. Third, all of the HCV sequences from the GenBank database were obtained by Sanger sequencing, which cannot detect the presence of RASs as minor variants, compared to the deep sequencing approaches (20% vs 1% cutoff).34 This methodology limits our study, especially in the context of retreatment strategies.
In conclusion, in this study, we evaluated whether RASs could be increased or coselected as a result of P/R treatments in HCV GT1a patients. Our results suggest that previous P/R treatment failure was not favorably associated with the increase in DAAs RAS that naturally exists in GT1a patients. Our results support the AASLD recommendations of DAA regimens for P/R treatment-experienced GT1a patients.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (30930082, 81171561 and 30972584) and the National Science and Technology Major Project of China (2008ZX10002-006, 2012ZX10002007001, 2017ZX10202203-007 and 2017ZX10202203-008).
Author contributions
PH and HR conceived the study, ZWC and XCP retrieved the data, ZWC, XCP and ZL analyzed the data, and all authors contributed to the writing of the manuscript. All authors also contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17(2):107–115. | ||
Wyles DL. Moving beyond interferon alfa: investigational drugs for hepatitis C virus infection. Top HIV Med. 2010;18(4):132–136. | ||
Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347(13):975–982. | ||
Vermehren J, Sarrazin C. New HCV therapies on the horizon. Clin Microbiol Infect. 2011;17(2):122–134. | ||
Zeuzem S. Clinical implications of hepatitis C viral kinetics. J Hepatol. 1999;31(Suppl 1):61–64. | ||
Dahl G, Sandstrom A, Akerblom E, Danielson UH. Resistance profiling of hepatitis C virus protease inhibitors using full-length NS3. Antivir Ther. 2007;12(5):733–740. | ||
Flint M, Mullen S, Deatly AM, et al. Selection and characterization of hepatitis C virus replicons dually resistant to the polymerase and protease inhibitors HCV-796 and boceprevir (SCH 503034). Antimicrob Agents Chemother. 2009;53(2):401–411. | ||
Strahotin CS, Babich M. Hepatitis C variability, patterns of resistance, and impact on therapy. Adv Virol. 2012;2012:267483. | ||
Contreras AM, Hiasa Y, He W, Terella A, Schmidt EV, Chung RT. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J Virol. 2002;76(17):8505–8517. | ||
Lontok E, Harrington P, Howe A, et al. Hepatitis C virus drug resistance-associated substitutions: state of the art summary. Hepatology. 2015;62(5):1623–1632. | ||
Liu R, Curry S, McMonagle P, et al. Susceptibilities of genotype 1a, 1b, and 3 hepatitis C virus variants to the NS5A inhibitor elbasvir. Antimicrob Agents Chemother. 2015;59(11):6922–6929. | ||
Forns X, Gordon SC, Zuckerman E, et al. Grazoprevir and elbasvir plus ribavirin for chronic HCV genotype-1 infection after failure of combination therapy containing a direct-acting antiviral agent. J Hepatol. 2015;63(3):564–572. | ||
Zeuzem S, Ghalib R, Reddy KR, et al. Grazoprevir-Elbasvir combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection: a randomized trial. Ann Intern Med. 2015;163(1):1–13. | ||
Sulkowski MS, Eron JJ, Wyles D, et al. Ombitasvir, paritaprevir co-dosed with ritonavir, dasabuvir, and ribavirin for hepatitis C in patients co-infected with HIV-1: a randomized trial. JAMA. 2015;313(12):1223–1231. | ||
Lawitz EJ, Dvory-Sobol H, Doehle BP, et al. Clinical Resistance to Velpatasvir (GS-5816), a novel pan-genotypic inhibitor of the hepatitis C virus NS5A protein. Antimicrob Agents Chemother. 2016;60(9):5368–5378. | ||
McPhee F, Hernandez D, Yu F, et al. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology. 2013;58(3):902–911. | ||
Howe AY, Black S, Curry S, et al. Virologic resistance analysis from a phase 2 study of MK-5172 combined with pegylated interferon/ribavirin in treatment-naive patients with hepatitis C virus genotype 1 infection. Clin Infect Dis. 2014;59(12):1657–1665. | ||
McPhee F, Sheaffer AK, Friborg J, et al. Preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-6,50,032). Antimicrob Agents Chemother. 2012;56(10):5387–5396. | ||
Jensen D, Sherman KE, Hezode C, et al. Daclatasvir and asunaprevir plus peginterferon alfa and ribavirin in HCV genotype 1 or 4 non-responders. J Hepatol. 2015;63(1):30–37. | ||
Pilot-Matias T, Tripathi R, Cohen D, et al. In vitro and in vivo antiviral activity and resistance profile of the hepatitis C virus NS3/4A protease inhibitor ABT-450. Antimicrob Agents Chemother. 2015;59(2):988–997. | ||
Coburn CA, Meinke PT, Chang W, et al. Discovery of MK-8742: an HCV NS5A inhibitor with broad genotype activity. ChemMedChem. 2013;8(12):1930–1940. | ||
Chen ZW, Li H, Ren H, Hu P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep. 2016;6:20310. | ||
EASL recommendations on treatment of hepatitis C 2015. J Hepatol. 2015;63(1):199–236. | ||
AASLD. HCV Guidance: Recommendations for Testing, Managing, and Treating Hepatitis C. 2017. Available from: http://www.hcvguidelines.org Accessed June 15, 2017. | ||
Sarrazin C, Lathouwers E, Peeters M, et al. Prevalence of the hepatitis C virus NS3 polymorphism Q80K in genotype 1 patients in the European region. Antiviral Res. 2015;116:10–16. | ||
Sarrazin C, Kieffer TL, Bartels D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132(5):1767–1777. | ||
McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. New Engl J Med. 2009;360(18):1827–1838. | ||
Kieffer TL, Sarrazin C, Miller JS, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46(3):631–639. | ||
Zeuzem S, Dusheiko GM, Salupere R, et al. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. New Engl J Med. 2014;370(21):1993–2001. | ||
Sede MM, Laufer NL, Quarleri J. Previous failure of interferon-based therapy does not alter the frequency of HCV NS3 protease or NS5B polymerase inhibitor resistance-associated variants: longitudinal analysis in HCV/HIV co-infected patients. Int J Antimicrob Agents. 2015;46(2):219–224. | ||
Lawitz E, Sulkowski MS, Ghalib R, et al. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: the COSMOS randomized study. Lancet. 2014;384(9956):1756–1765. | ||
Afdhal N, Reddy KR, Nelson DR, et al. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med. 2014;370(16):1483–1493. | ||
Zeuzem S, Jacobson IM, Baykal T, et al. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med. 2014;370(17):1604–1614. | ||
Beloukas A, King S, Childs K, et al. Detection of the NS3 Q80K polymorphism by Sanger and deep sequencing in hepatitis C virus genotype 1a strains in the UK. Clin Microb. 2015;21(11):1033–1039. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.