Back to Journals » International Journal of General Medicine » Volume 14
Patterns of Immune Infiltration and the Key Immune-Related Genes in Acute Type A Aortic Dissection in Bioinformatics Analyses
Received 25 April 2021
Accepted for publication 14 June 2021
Published 25 June 2021 Volume 2021:14 Pages 2857—2869
DOI https://doi.org/10.2147/IJGM.S317405
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Fengshou Chen, Jie Han, Bing Tang
Department of Anesthesiology, The First Hospital of China Medical University, Shenyang, Liaoning Province, People’s Republic of China
Correspondence: Bing Tang No. 155 Nangjing North Street, Shenyang, Liaoning Province, People’s Republic of China
Tel +86 24 83283100
Email [email protected]
Background: Immune-inflammatory mechanisms contribute greatly to the complex process leading to type A aortic dissection (TAAD). This study aims to explore immune infiltration and key immune-related genes in acute TAAD.
Methods: ImmuCellAI algorithm was applied to analyze patterns of immune infiltration in TAAD samples and normal aortic vessel samples in the GSE153434 dataset. Differentially expressed genes (DEGs) were screened. Immune-related genes were obtained from overlapping DEGs of GSE153434 and immune genes of the ImmPort database. The hub genes were obtained based on the protein–protein interaction (PPI) network. The hub genes in TAAD were validated in the GSE52093 dataset. The correlation between the key immune-related genes and infiltrating immune cells was further analyzed.
Results: In the study, the abundance of macrophages, neutrophils, natural killer T cells (NKT cells), natural regulatory T cells (nTreg), T-helper 17 cells (Th17 cells) and monocytes was increased in TAAD samples, whereas that of dendritic cells (DCs), CD4 T cells, central memory T cells (Tcm), mucosa associated invariant T cells (MAIT cells) and B cells was decreased. Interleukin 6 (IL-6), C-C motif chemokine ligand 2 (CCL2) and hepatocyte growth factor (HGF) were identified and validated in the GSE52093 dataset as the key immune-related genes. Furthermore, IL-6, CCL2 and HGF were correlated with different types of immune cells.
Conclusion: In conclusion, several immune cells such as macrophages, neutrophils, NKT cells, and nTreg may be involved in the development of TAAD. IL-6, CCL2 and HGF were identified and validated as the key immune-related genes of TAAD via bioinformatics analyses. The key immune cells and immune-related genes have the potential to be developed as targets of prevention and immunotherapy for patients with TAAD.
Keywords: acute type A aortic dissection, immune infiltration, ImmuCellAI, bioinformatics
Introduction
Aortic dissection (AD) is a rare life-threatening condition in the aorta with high mortality and morbidity.1 The mortality rate of acute type A aortic dissection (TAAD), the most severe form of AD, reaches 26–58% in patients who have undergone surgery or been treated noninvasively.2 TAAD is a surgical emergency and its management has the complexity and poor outcomes when there is organ malperfusion.3
Many pathological processes including dyslipidemia, smoking, hypertension and vascular inflammation, interact with each other and accelerate the evolution of TAAD.1 Particular autoimmune and infectious diseases are implicated in the development of AD. Persistent aortic inflammation and changes in the degree of elasticity of the aortic media greatly increase the risk of intimal disruption.4 Recently, it has been shown that immune-inflammatory mechanisms contribute greatly to aortic wall remodeling, suggesting that immune response are common steps in the complex process leading to TAAD.5
This study aims to analyze the infiltration of immune cells in TAAD and explore key immune-related genes in TAAD. The TAAD dataset GSE1534346 was downloaded from the Gene Expression Omnibus (GEO) database. Immune Cell Abundance Identifier (ImmuCellAI)7 algorithm was used to quantify the proportions of immune cells in TAAD samples and normal tissues. Differentially expressed genes (DEGs) of the TAAD and normal tissues were analyzed. The identified key immune-related genes were validated in another TAAD dataset GSE52093.8 The relationship between these genes and infiltrating immune cells was also identified.
Methods
Data Acquisition and Processing
Two gene expression datasets, GSE153434 and GSE52093, were downloaded from the GEO database. GSE153434 (including 10 TAAD samples and 10 normal aortic vessel samples) was used to analyze DEGs and immune infiltration in TAAD. Limma and SVA packages in R were used to correct the sample data.9 In addition, the GSE52093 dataset (including 7 TAAD samples and 5 normal aortic vessel samples) was used as the validation cohort.
Analysis of DEGs
The data were normalized and transformed into a log2-based logarithm using the robust multiarray average algorithm after background correction.10–12 Thereafter, the internal standard probe was removed and a box plot was drawn for the expression values of every chip before and after normalization. In addition, the limma package in R was used to analyze four subsets of DEGs,13 genes with a |log2 FC| > 1 and p < 0.05.
Patterns of Immune Infiltration in TAAD
ImmuCellAI (http://bioinfo.life.hust.edu.cn/web/ImmuCellAI/)7 algorithm was used to estimate the abundance of 24 types of immune cells based on a gene expression data set.14 Eighteen T-cell subsets and six key immune cells including natural killer cells (NK cells), dendritic cells (DCs), neutrophils, monocytes, macrophages and B cells.
Principal Component Analysis (PCA)
PCA is a linear transform that extracts the essential elements in the data using a covariance matrix or a correlation matrix and can reduce the analysis dimension and determine the essential elements.15 We performed PCA to realine the dimensionality of the 24 types of immune infiltrating cells while retaining key information.16
Expression Profiles of Immune-Related Genes
The list of immune-related genes was download from the ImmPort database (www.immport.org).17 Differentially expressed immune-related genes were intersected with immune-related genes and DEGs in GSE52093.
Enrichment Analysis for the DEGs
The Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on Metascape, a powerful annotation tool for the analysis of gene function.18,19 Terms with a p value <0.05, a minimum count of 3 and an enrichment factor >1.5 were collected and grouped into clusters based on their similarities in membership (Kappa scores >0.3).20,21 Thereafter, the top 20 terms for GO or pathway enrichment terms were selected for presentation.
Construction of a Protein-Protein Interaction (PPI) Network
A PPI network for the enriched DEGs from KEGG analysis was constructed using the STRING database and an interaction score with a median confidence of 0.4 was set as the standard cutoff criterion.12,22 The network was constructed using the Cytoscape software platform based on the information of the functional analysis.23 Additionally, the hub genes were identified via cytoHubba in Cytoscape. The maximum neighborhood component (MNC), edge percolated component (EPC), node connect degree (Degree) and closeness algorithms were used for selecting hub genes from PPI networks in the cytoHubba plugin.24–26
Correlation Analysis Between Infiltrating Immune Cells and Identified Genes
Spearman’s rank correlation analysis in R software was used to explore the association of the hub genes with infiltrating immune cells. The “ggplot2” package was applied for the charting to demonstrate the results.
Statistical Analysis
The differences of gene expression and immune cell infiltration were tested by Wilcoxon rank-sum test, and Pearson correlation coefficients were calculated to reveal the correlation between any two immune infiltrating cells. The relationship between the expression of gene biomarkers and infiltrating immune cells was analyzed using Spearman correlation coefficient. DEGs between TAAD and normal aorta were analyzed with the volcano chart by Limma package in R, and heatmap was drawn by “heatmap” package in R.
Results
Immune Infiltration Analysis
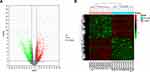
ImmuCellAI algorithm was used to analyze immune cell infiltration in 10 TAAD samples and 10 normal aortic samples and a significant difference in the abundance of different types of immune cells was found (Figure 1A and Table 1). PCA analysis results show that TAAD samples and normal aortic samples could be significantly distinguished (Figure 1B). The abundance of T-helper 17 cells (Th17 cells), neutrophils, T-helper 2 cells (Th2 cells), mucosa associated invariant T cells (MAIT cells), and macrophages show significant differences in TAAD samples and normal aortic samples (Figure 1C).
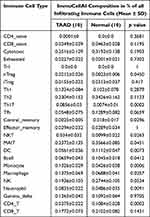 |
Table 1 Comparison of ImmuCellAI Immune Cell Compositions Between TAAD and Normal Tissues |
In addition, the results reveal a correlation map displaying the Pearson correlation values for each comparison between the immune cells. B cells have a positive correlation value of 0.35 with CD4 T cells, 0.49 with central memory T cells (Tcm), 0.05 with macrophages, 0.19 with MAIT cells, 0.02 with neutrophils, 0.75 with dendritic cells (DCs). However, B cells have a negative correlation value of −0.44 with natural killer T cells (NKT cells), −0.32 with natural regulatory T cells (nTreg), and −0.5 with Th17, −0.43 with monocytes (Figure 2).
 |
Figure 2 Correlation among all immune cells. |
The results clearly demonstrate that there are 11 types of immune cells with different abundance in TAAD samples and normal aortic samples. The abundance of macrophages (p = 0.026), neutrophils (p = 0.0091), NKT cells (p = 0.026), nTreg (p = 0.045), Th17 (p = 0.00016) and monocytes (p = 0.00058) is increased in TAAD samples (Figure 3), whereas that DCs (p = 0.0073), CD4 T cells (p = 0.00033), Tcm (p = 0.03), MAIT cells (p = 0.045) and B cells (p = 0.041) is decreased in TAAD samples (Figure 4).
Screening of Differentially Expressed Immune-Related Genes
DEGs in the GSE153434 dataset including 10 TAAD samples and 10 normal aortic samples were analyzed in the study. A total of 2286 DEGs in the TAAD samples were identified. The volcano chart and heatmap demonstrating the gene expression profiles of DEGs are presented in Figures 5A and B. The detailed information about DEGs is listed in Supplementary Table S1. Furthermore, we obtained 228 overlapping immune-related genes of DEGs in GSE153434 and immune-related genes from ImmPort as shown in Supplementary Table S2.
GO and KEGG Enrichment Analyses
The GO and KEGG analysis of 228 immune-related genes were performed by DAVID. The immune-related genes are mostly enriched in biological processes (BP) including immune response, inflammatory response, positive regulation of ERK1 and ERK2 cascade, intrinsic apoptotic signaling pathway, regulation of MAP kinase activity and chemokine-mediated signaling pathway, as shown in Figure 6A. The outcomes of CC and MF enrichment analysis are listed in Figures 6B and C with specific items. In particular, the immune-related genes are mostly enriched in the KEGG pathway including Cytokine-cytokine receptor interaction, Ras signaling pathway, TNF signaling pathway, Chemokine signaling pathway and PI3K-Akt signaling pathway, as shown in Figure 6D.
Construction and Analysis of the PPI Network
The PPI network of 228 immune-related genes was constructed to distinguish the hub DEGs from the common DEGs (Figure 7). Key immune-related genes were selected using cytoHubba. Four hub genes were identified by the overlap of the top five immune-related genes according to four ranked methods in cytoHubba (Table 2). The four key immune-related genes are interleukin 6 (IL-6), vascular endothelial growth factor A (VEGFA), C-C motif chemokine ligand 2 (CCL2) and hepatocyte growth factor (HGF).
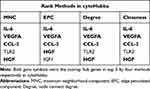 |
Table 2 Hub Genes for TAAD Ranked in cytoHubba |
 |
Figure 7 The protein-protein interaction network of DEGs between TAAD tissues and normal tissues. Red circular nodes were identified as hub genes. |
Validation of Key Immune-Related Genes in GSE52093
Furthermore, the GSE52093 dataset was used to validate the expression levels of the four key immune-related genes. The expression levels of IL-6, CCL2 and HGF in TAAD samples are significantly higher than those in the normal aortic samples (Table 3; all p < 0.05). However, the expression levels of VEGFA in TAAD samples and the normal aortic samples show no significant difference (Table 3). Therefore, IL-6, CCL2 and HGF were identified as key immune-related genes in TAAD.
 |
Table 3 Four Hub DEGs in GSE153434 and GSE52093 Datasets |
Correlation Analysis Between the Key Immune-Related Genes and Immune Infiltration Cells
As shown in Figure 8A, IL-6 is positively correlated with Gamma delta T cells (γδ T cells) (r = 0.49, p = 0.0276) and nTreg (r = 0.53, p = 0.0167). CCL-2 is positively correlated with monocytes (r = 0.49, p = 0.0276), neutrophils (r = 0.52, p = 0.0172), Th17 (r = 0.64, p = 0.0023) and nTreg (r = 0.66, p = 0.0015) and negatively correlated with CD4 T cells (r = −0.63, p = 0.0030), DC cells (r = −0.54, p = 0.0133), Tcm (r = −0.51, p = 0.0201) and MAIT cells (r = −0.46, p = 0.0424), as shown in Figure 8B. HGF is positively correlated with monocytes (r = 0.55, p = 0.0111), Th17 (r = 0.58, p = 0.0073) and macrophages (r = 0.55, p = 0.0123) and negatively correlated with CD4 T cells (r = −0.69, p = 0.0008) and Tfh cells (r = −0.50, p = 0.0255; Figure 8C).
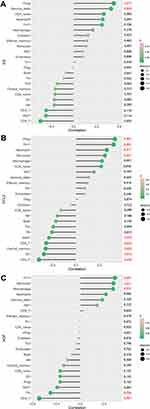 |
Figure 8 Correlation between IL-6 (A), CCL2 (B), HGF (C), and infiltrating immune cells in TAAD. |
Discussion
In this study, the GSE153434 dataset including 20 samples was used to analyze the infiltration of immune cells in TAAD and the correlation among immune cells. Moreover, the genes that play a key role in TAAD were screened out by constructing a network and the results were validated in the GSE52093 dataset, making the results more reliable.
In this study, the infiltration ratio of 24 types of immune cells in TAAD samples and normal aortic samples shows a significant difference. PCA analysis demonstrates that based on the infiltration of immune cells, TAAD samples and normal aortic samples could be distinguished. The abundance of macrophages, neutrophils, NKT cells, nTreg, Th17 and monocytes is increased in TAAD samples, while that of DC cells, B cells, Tcm, MAIT cells and CD4 T cells is decreased in TAAD samples, suggesting that several types of immune cells are involved in the AD, including TAAD. Macrophages, which have both anti-inflammatory and inflammatory effects, are related to the pathophysiology mechanism of AD.27 Macrophages play multiple roles in TAAD and the release of VEGF by macrophages is positive in the tunica media, and all AAD samples show immune-inflammatory infiltration and vessel growth.28 As the dominant inflammatory cells in acute inflammation, neutrophils mainly infiltrate the adventitia of AD tissue.29 Neutrophil to lymphocyte ratio (NLR) is markedly increased in patients with TAAD and is a prognostic predictor for TAAD.30 As a specialized lymphocyte subset recognizing lipid antigens, NKT cells can be pro- or anti-inflammatory.31 NKT cells in the peripheral blood of patients with TAAD are significantly increased.32 Aortic atherosclerosis is more associated with aortic dissection.33 Atherosclerotic plaques in the aortic root of mice treated with Thymic stromal lymphopoietin (TSLP) and TSLP-conditioned bone marrow dendritic cells (TSLP-DCs) are fewer than those in the control group. Besides, the number of nTregs and CD4+LAP+ Tregs in the spleen is increased.34 Th17 is implicated in vascular pathology.35 In patients with thoracic aortic dissections, Th17 infiltration is increased. Monocytes can be classified into nonclassical monocytes, intermediate monocytes, and classical monocytes with distinctive functional and phenotypic properties. A previous study has revealed that intermediate monocytes are notably decreased in the TAAD group versus carotid artery stenosis and traditional cardiovascular risk factors groups, whereas classical monocytes are notably increased in the TAAD group versus carotid artery stenosis group.36 As key immune modulators, DCs can mount immune tolerance or responses.37 The normal intima is thought to be populated by vascular DCs that, during hypercholesterolemia, initiate atherosclerosis by being the first to accumulate cholesterol.38 TLR7 stimulation is associated with the expansion of B-cell and Treg response, which may have systemic and local effects on lesions to prevent arterial lipid accumulation and inflammation.39 B cells might participate in the pathophysiology of β-aminopropionitrile-induced TAD in mice.40 Memory T cells play an important role to regulate chronic inflammation and delay the progression of the pathogenesis of atherosclerosis via the AMPK signaling pathway.41 MAIT cells, which might be involved with vascular dysfunction have been involved in various inflammatory and autoimmune diseases.42,43 The results in this study conform to these findings. The roles of these immune cells in TAAD still need further study.
A total of 228 immune-related genes of DEGs in GSE153434 and immune genes from ImmPort were identified in this study. The GO and KEGG analysis of these immune-related genes were performed by DAVID. The immune-related genes were mostly enriched in biological processes (BP) including inflammatory response and immune response. In particular, the immune-related genes were mostly enriched in the KEGG pathway including TNF signaling pathway, Cytokine-cytokine receptor interaction and Chemokine signaling pathway. Obtained biological processes and pathways were closely associated with inflammation and immunity. Regulation of inflammatory response was identified as one important pathway involved in the pathological progress of TAAD.44 Previous studies have identified the involvement of chemokine and cytokine. A significant increase in CCL2, TNF-α, IL-10 and IL-8 was observed in the peripheral blood of patients with TAAD.32 Local neutrophil recruitment and activation were triggered by adventitial CXCL1/granulocyte-colony stimulating factor expression in response to AAD.45
Four hub immune-related genes in TAAD were identified from the PPI network, including IL-6, CCL2, VEGFA and HGF using cytoHubba in Cytoscape. Moreover, the GSE52093 dataset was used to validate the expression of four key immune-related genes and it was found that the trend of IL-6, CCL2 and HGF was similar to that in the GSE153434 dataset. Vascular inflammation is a common pathophysiological response to aortic aneurysms/dissection and the NF-kappaB/IL-6 signaling pathway mediating vascular inflammation.46 IL-6 level in the TAAD group significantly increased compared to those in the control group.28,47 D-dimer > 14.0 μg/mL and IL-6 > 108 pg/mL can be the best predictive signs for early poor postoperative prognosis after TAAD.48 Tissue homogenized levels of IL-6 and TNF-α markedly increased in the TAAD group compared with those in the healthy group.49 The mRNA expression level of CCL2 notably increased in the mice AD model group compared with that in the control group.50 Serum CCL2 level in the AAD group was also significantly higher than that in the control group.51 CCL2 plays a key role in atherosclerosis and the monocyte containing MCPs regulates the growth of other types of cells within the atherosclerotic lesion.52 The interaction between macrophages and aortic adventitial fibroblasts provides an IL-6/CCL2 amplification loop for the acceleration of vascular inflammation leading to aortic dissection in mice.53 HGF is a pleiotropic cytokine involved in multiple biological and cellular processes, including the improvement of alleviation of inflammation and cell injury.54 In patients with AD, higher HGF concentrations in serum have been reported to be correlated with the thrombi.55 Furthermore, correlation analysis between immune cells and IL-6, CCL2, HGF was performed respectively. IL-6 and CCL2 were found to be positively correlated with nTreg. CCL2 and HGF were found to be positively correlated with monocytes and Th17 was positively correlated with CD4 T cells. The existing evidence and our findings reveal that several types of infiltrating immune cells and three key immune-related genes play important roles in TAAD and deserve future exploration.
While the study uncovered some insightful findings, it had several limitations. First, the results and the validation process were based on bioinformatics analysis. mRNA expression levels in the microarray analysis may not be directly correlated with the protein expression levels, and the calculated value of immune cells may not directly show the abundance of infiltrating immune cells in the tissue. Second, the sample size in the GSE153434 and GSE52093 datasets was relatively small. In addition, clinical information concerning the GSE153434 and GSE52093 datasets was not available. Therefore, the validation of the results at the cell and protein levels using different methods such as immunoblotting and immunostaining might be better. A multicentre study with larger sample size is still needed in the future.
In conclusion, IL-6, CCL2 and HGF were identified and validated as the key immune-related genes of TAAD via bioinformatics analyses. Macrophages, neutrophils, NKT cells, nTreg, Th17, monocytes, DCs, CD4 T cells, Tcm, MAIT cells and B cells might be involved in the pathogenesis of TAAD. These key immune-related genes and immune cells might help to further provide novel insights for candidate targets of prevention and immunotherapy for patients with TAAD.
Data Sharing Statement
The GSE1534346 and GSE520938 dataset was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/). Other datasets supporting the conclusions in this article are included within the article and its additional files.
Acknowledgments
We would like to acknowledge the reviewers for their invaluable comments on this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest and are solely responsible for the content and writing of this manuscript.
References
1. Gudbjartsson T, Ahlsson A, Geirsson A, et al. Acute type A aortic dissection - a review. Scand Cardiovasc J: SCJ. 2020;54(1):1–13. doi:10.1080/14017431.2019.1660401
2. Jiang T, Si L. Identification of the molecular mechanisms associated with acute type A aortic dissection through bioinformatics methods. Braz J Med Biol Res. 2019;52(11):e8950. doi:10.1590/1414-431x20198950
3. Munir W, Chong JH, Harky A, Bashir M, Adams B. Type A aortic dissection: involvement of carotid artery and impact on cerebral malperfusion. Asian Cardiovasc Thorac Ann. 2020;218492320984329.
4. Elsayed RS, Cohen RG, Fleischman F, Bowdish ME. Acute type A aortic dissection. Cardiol Clin. 2017;35(3):331–345. doi:10.1016/j.ccl.2017.03.004
5. Cifani N, Proietta M, Tritapepe L, et al. Stanford-A acute aortic dissection, inflammation, and metalloproteinases: a review. Ann Med. 2015;47(6):441–446. doi:10.3109/07853890.2015.1073346
6. Zhou Z, Liu Y, Zhu X, et al. Exaggerated autophagy in stanford type A aortic dissection: a transcriptome pilot analysis of human ascending aortic tissues. Genes. 2020;11(10):1187. doi:10.3390/genes11101187
7. Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci. 2020;7(7):1902880. doi:10.1002/advs.201902880
8. Pan S, Wu D, Teschendorff AE, et al. JAK2-centered interactome hotspot identified by an integrative network algorithm in acute Stanford type A aortic dissection. PLoS One. 2014;9(2):e89406. doi:10.1371/journal.pone.0089406
9. Nie H, Qiu J, Wen S, Zhou W. Combining bioinformatics techniques to study the key immune-related genes in abdominal aortic aneurysm. Front Genet. 2020;11:579215. doi:10.3389/fgene.2020.579215
10. Chen H, Chen C, Yuan X, et al. Identification of immune cell landscape and construction of a novel diagnostic nomogram for Crohn’s disease. Front Genet. 2020;11:423. doi:10.3389/fgene.2020.00423
11. Xu WH, Wu J, Wang J, et al. Screening and identification of potential prognostic biomarkers in adrenocortical carcinoma. Front Genet. 2019;10:821. doi:10.3389/fgene.2019.00821
12. Ma L, Lu H, Chen R, et al. Identification of key genes and potential new biomarkers for ovarian aging: a study based on RNA-sequencing data. Front Genet. 2020;11:590660. doi:10.3389/fgene.2020.590660
13. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
14. Mei J, Yang X, Xia D, et al. Systematic summarization of the expression profiles and prognostic roles of the dishevelled gene family in hepatocellular carcinoma. Mol Genet Genom Med. 2020;8(9):e1384. doi:10.1002/mgg3.1384
15. Lei C, Yang D, Chen S, et al. Patterns of immune infiltration in stable and raptured abdominal aortic aneurysms: a gene-expression-based retrospective study. Gene. 2020;762:145056. doi:10.1016/j.gene.2020.145056
16. Nie H, Wang S, Wu Q, Xue D, Zhou W. Five immune-gene-signatures participate in the development and pathogenesis of Kawasaki disease. Immun Inflamm Dis. 2020.
17. Bhattacharya S, Andorf S, Gomes L, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2–3):234–239. doi:10.1007/s12026-014-8516-1
18. Zhang YF, Meng LB, Hao ML, Yang JF, Zou T. Identification of co-expressed genes between atrial fibrillation and stroke. Front Neurol. 2020;11:184. doi:10.3389/fneur.2020.00184
19. Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33(10):111. doi:10.1007/s12032-016-0829-6
20. Niu X, Zhang J, Zhang L, et al. Weighted gene co-expression network analysis identifies critical genes in the development of heart failure after acute myocardial infarction. Front Genet. 2019;10:1214. doi:10.3389/fgene.2019.01214
21. Qi L, Liu B, Chen X, et al. Single-cell transcriptomic analysis reveals mitochondrial dynamics in oocytes of patients with polycystic ovary syndrome. Front Genet. 2020;11:396. doi:10.3389/fgene.2020.00396
22. Kumari N, Karmakar A, Chakrabarti S, Ganesan SK. Integrative computational approach revealed crucial genes associated with different stages of diabetic retinopathy. Front Genet. 2020;11:576442. doi:10.3389/fgene.2020.576442
23. Chen L, Zhang S, Zhang S, et al. Identification of long non-coding RNA-associated competing endogenous RNA network in the differentiation of chicken preadipocytes. Genes. 2019;10(10):795. doi:10.3390/genes10100795
24. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
25. Ma Z, Xu J, Ru L, Zhu W. Identification of pivotal genes associated with the prognosis of gastric carcinoma through integrated analysis. Biosci Rep. 2021;41(4). doi:10.1042/BSR20203676
26. Chaudhary R, Balhara M, Jangir DK, Dangi M, Dangi M, Chhillar AK. In silico protein interaction network analysis of virulence proteins associated with invasive aspergillosis for drug discovery. Curr Top Med Chem. 2019;19(2):146–155. doi:10.2174/1568026619666181120150633
27. Wang X, Zhang H, Cao L, He Y, Ma A, Guo W. The role of macrophages in aortic dissection. Front Physiol. 2020;11:54. doi:10.3389/fphys.2020.00054
28. Del Porto F, Di Gioia C, Tritapepe L, et al. The multitasking role of macrophages in Stanford type A acute aortic dissection. Cardiology. 2014;127(2):123–129. doi:10.1159/000355253
29. Yoshida S, Yamamoto M, Aoki H, et al. STAT3 activation correlates with adventitial neutrophil infiltration in human aortic dissection. Ann Vasc Dis. 2019;12(2):187–193. doi:10.3400/avd.oa.19-00007
30. Bedel C, Selvi F. Association of platelet to lymphocyte and neutrophil to lymphocyte ratios with in-hospital mortality in patients with type A acute aortic dissection. Braz J Cardiovasc Surg. 2019;34(6):694–698. doi:10.21470/1678-9741-2018-0343
31. Subramanian S, Turner MS, Ding Y, et al. Increased levels of invariant natural killer T lymphocytes worsen metabolic abnormalities and atherosclerosis in obese mice. J Lipid Res. 2013;54(10):2831–2841. doi:10.1194/jlr.M041020
32. Del Porto F, Proietta M, Tritapepe L, et al. Inflammation and immune response in acute aortic dissection. Ann Med. 2010;42(8):622–629. doi:10.3109/07853890.2010.518156
33. Barbetseas J, Alexopoulos N, Brili S, et al. Atherosclerosis of the aorta in patients with acute thoracic aortic dissection. Circulat J. 2008;72(11):1773–1776. doi:10.1253/circj.CJ-08-0433
34. Yu K, Zhu P, Dong Q, et al. Thymic stromal lymphopoietin attenuates the development of atherosclerosis in ApoE-/- mice. J Am Heart Assoc. 2013;2(5):e000391. doi:10.1161/JAHA.113.000391
35. Ju X, Ijaz T, Sun H, et al. Interleukin-6-signal transducer and activator of transcription-3 signaling mediates aortic dissections induced by angiotensin II via the T-helper lymphocyte 17-interleukin 17 axis in C57BL/6 mice. Arterioscler Thromb Vasc Biol. 2013;33(7):1612–1621. doi:10.1161/ATVBAHA.112.301049
36. Cifani N, Proietta M, Taurino M, Tritapepe L, Del Porto F. Monocyte subsets, stanford-a acute aortic dissection, and carotid artery stenosis: new evidences. J Immunol Res. 2019;2019:9782594. doi:10.1155/2019/9782594
37. Scheenstra MR, Martínez-Botía P, Acebes-Huerta A, et al. Comparison of the PU.1 transcriptional regulome and interactome in human and mouse inflammatory dendritic cells. J Leukoc Biol. 2020. doi:10.1002/JLB.6A1219-711RRR
38. Williams JW, Zaitsev K, Kim KW, et al. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nat Immunol. 2020;21(10):1194–1204. doi:10.1038/s41590-020-0768-4
39. Karadimou G, Gisterå A, Gallina AL, et al. Treatment with a Toll-like Receptor 7 ligand evokes protective immunity against atherosclerosis in hypercholesterolaemic mice. J Intern Med. 2020;288(3):321–334. doi:10.1111/joim.13085
40. Gao Y, Wang Z, Zhao J, et al. Involvement of B cells in the pathophysiology of β-aminopropionitrile-induced thoracic aortic dissection in mice. Exp Animals. 2019;68(3):331–339. doi:10.1538/expanim.18-0170
41. Peng LP, Cao Y, Zhao SL, Huang YX, Yang K, Huang W. Memory T cells delay the progression of atherosclerosis via AMPK signaling pathway. Ann Transl Med. 2019;7(23):782. doi:10.21037/atm.2019.11.20
42. Fazekas B, Moreno-Olivera A, Kelly Y, et al. Alterations in circulating lymphoid cell populations in systemic small vessel vasculitis are non-specific manifestations of renal injury. Clin Exp Immunol. 2018;191(2):180–188. doi:10.1111/cei.13058
43. Emgård J, Bergsten H, McCormick JK, et al. MAIT cells are major contributors to the cytokine response in group A streptococcal toxic shock syndrome. Proc Natl Acad Sci U S A. 2019;116(51):25923–25931. doi:10.1073/pnas.1910883116
44. Kimura N, Futamura K, Arakawa M, et al. Gene expression profiling of acute type A aortic dissection combined with in vitro assessment. Eur J Cardio-Thorac Surg. 2017;52(4):810–817. doi:10.1093/ejcts/ezx095
45. Anzai A, Shimoda M, Endo J, et al. Adventitial CXCL1/G-CSF expression in response to acute aortic dissection triggers local neutrophil recruitment and activation leading to aortic rupture. Circ Res. 2015;116(4):612–623. doi:10.1161/CIRCRESAHA.116.304918
46. Brasier AR. The nuclear factor-kappaB-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc Res. 2010;86(2):211–218. doi:10.1093/cvr/cvq076
47. Liu H, Luo Z, Liu L, et al. Inflammatory biomarkers to predict adverse outcomes in postoperative patients with acute type A aortic dissection. Scand Cardiovasc J: SCJ. 2020;54(1):37–46. doi:10.1080/14017431.2019.1689289
48. Wu Q, Li J, Chen L, et al. Efficacy of interleukin-6 in combination with D-dimer in predicting early poor postoperative prognosis after acute stanford type a aortic dissection. J Cardiothorac Surg. 2020;15(1):172. doi:10.1186/s13019-020-01206-y
49. Bai Z, Gu J, Shi Y, Meng W. Effect of inflammation on the biomechanical strength of involved aorta in type A aortic dissection and ascending thoracic aortic aneurysm: an initial research. Anatol J Cardiol. 2018;20(2):85–92. doi:10.14744/AnatolJCardiol.2018.49344
50. Zhao JQ, Gao YX, Wu C, et al. [Effects of alprostadil in β-aminopropanitrile induced aortic dissection in a murine model]. Zhonghua Xin xue Guan Bing Za Zhi. 2020;48(8):682–688. [Chinese]. doi:10.3760/cma.j.cn112148-20190925-00592
51. Luo FY, Liu ZH, Jiang HH, Lin GQ. Correlation between plasma level of monocyte chemotactic protein 1 and acute aortic dissection. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2015;37(3):352–354. doi:10.3881/j.issn.1000-503X.2015.03.021
52. Lin J, Kakkar V, Lu X. Impact of MCP-1 in atherosclerosis. Curr Pharm Des. 2014;20(28):4580–4588. doi:10.2174/1381612820666140522115801
53. Tieu BC, Lee C, Sun H, et al. An adventitial IL-6/MCP1 amplification loop accelerates macrophage-mediated vascular inflammation leading to aortic dissection in mice. J Clin Invest. 2009;119(12):3637–3651. doi:10.1172/JCI38308
54. Peng F, Chang W, Sun Q, et al. HGF alleviates septic endothelial injury by inhibiting pyroptosis via the mTOR signalling pathway. Respir Res. 2020;21(1):215. doi:10.1186/s12931-020-01480-3
55. Hata N, Matsumori A, Yokoyama S, et al. Hepatocyte growth factor and cardiovascular thrombosis in patients admitted to the intensive care unit. Circulat J. 2004;68(7):645–649. doi:10.1253/circj.68.645
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.