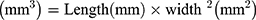Back to Journals » International Journal of Nanomedicine » Volume 15
Distinct Proteins in Protein Corona of Nanoparticles Represent a Promising Venue for Endogenous Targeting – Part II: In vitro and in vivo Kinetics Study
Authors Sebak AA , Gomaa IEO , ElMeshad AN , Farag MH, Breitinger U , Breitinger H , AbdelKader MH
Received 7 August 2020
Accepted for publication 30 October 2020
Published 30 November 2020 Volume 2020:15 Pages 9539—9556
DOI https://doi.org/10.2147/IJN.S273721
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Aya Ahmed Sebak,1 Iman Emam Omar Gomaa,2 Aliaa Nabil ElMeshad,3 Mahmoud Hussien Farag,1 Ulrike Breitinger,4 Hans-Georg Breitinger,4 Mahmoud Hashem AbdelKader5,6
1Pharmaceutical Technology Department, Faculty of Pharmacy and Biotechnology, German University in Cairo (GUC), New Cairo City, Egypt; 2Biochemistry Department, Faculty of Pharmacy, October University for Modern Sciences and Arts (MSA), Giza, Egypt; 3Department of Pharmaceutics and Industrial Pharmacy, Faculty of Pharmacy, Cairo University, Cairo, Egypt; 4Biochemistry Department, Faculty of Pharmacy and Biotechnology, German University in Cairo (GUC), New Cairo City, Egypt; 5National Institute of Laser Enhanced Sciences (NILES), Cairo University (CU), Giza, Egypt; 6European University in Egypt (EUE), New Administrative Capital, Cairo, Egypt
Correspondence: Aya Ahmed Sebak
Faculty of Pharmacy and Biotechnology, German University in Cairo (GUC), New Cairo City, Egypt
Tel +20 1205727787
Email [email protected]
Iman Emam Omar Gomaa
Biochemistry Department Faculty of Pharmacy, October University for Modern Sciences and Arts (MSA), Giza, Egypt
Tel +20 1275146432
Email [email protected]
Introduction: Nanoparticles (NPs), upon introduction to the biological systems, become wrapped by serum and cellular proteins constituting the protein corona (PC). This PC contributes largely to the NPs’ interaction with the biological systems and their subsequent functions. On the one hand, PC can decrease the efficiency of targeting by directing the NPs to the reticuloendothelial system (RES) or by masking the active targeting moieties and decreasing their ability to bind to their target receptors. On the other hand, some components of PC have offered hopes for achieving endogenous targeting.
Methods: In this study, we aimed at the investigation of the role of the PC in determining the behavior of cRGDyk peptide-unconjugated and -conjugated NPs (uNPs and cNPs) exhibiting different physicochemical properties and their interaction with melanoma on in vitro and in vivo levels. Mathematical modeling has been utilized to understand the kinetics of the interaction of NPs with the tumor cells and different organs, respectively.
Results: Endocytosis and exocytosis were reported to occur simultaneously for the utilized NPs. The balance was largely dependent on the NPs’ physicochemical properties and the role of the PC. In addition, distinct proteins present in the PC (illustrated in the results of the PC analysis in part I) have also determined the patterns of the NPs’ distribution in different organs and tissues of the vascular system, the RES system and the target tumot tissue. Vitronectin (VN) was found to mediate higher accumulation in integrin receptor-expressing melanoma cells, while complement 3 protein (C3) and clusterin (CLU), as an opsonin and dysopsonin, respectively, regulated the balance between the RES uptake and blood circulation.
Discussion: PC, if properly modulated by tuning NPs’ physicochemical properties, can serve as a potential venue for optimum utilization of NPs in cancer therapy.
Keywords: Protein corona, endogenous targeting, melanoma, endocytosis, exocytosis, biodistribution, pharmacokinetics
Introduction
Nanocarriers, in general, preferentially accumulate in tumor tissues by the enhanced permeability and retention (EPR) effect. The tumor possesses a leaky vasculature, which allows the extravasation of NPs from the blood capillaries. In addition, it lacks lymphatic drainage allowing the drug-loaded NPs to reside at the tumor site for a longer duration compared to the free drug molecules. However, overreliance on passive targeting is sometimes misleading; EPR effect is a heterogeneous phenomenon that is not a universal property of all types of cancer.1–4 Passive targeting can improve the contact probability between the NPs and the target cells; however, it does not affect the intracellular uptake of the NPs which is largely dependent on the carrier itself.5,6 For example, the particle shape has been reported to affect the intracellular uptake of the NPs and their biodistribution.7 Additionally, the particle size8,9 and the particle rigidity10,11 play important roles. Active targeting, which employs ligands with a specific affinity towards the target site, has long been regarded as the optimum strategy for maximizing the tumor accumulation of NPs.12,13 Active targeting ligands potentially increase the NPs’ accumulation in their target site by anchoring them onto the cells. This allows the NPs to be retained more efficiently and rapidly than NPs relying only on the passive targeting potential.3,4,14,15 However, NPs do not have any propulsive power that guides them towards their target, they eventually accumulate in the tumor tissue by virtue of the EPR effect. Therefore, the enhanced tumor uptake upon ligand conjugation is at the mercy of the blood circulation.3,4,6,13,15,16
The formation of PC on NPs’ surfaces upon the in vivo administration is inevitable. Moreover, it largely determines the NPs’ distribution, their tissue accumulation and their interaction with the target and non-target cells.17,18 The composition of PC has been regarded as a fingerprint for each formulation of NPs, which determines distinct downstream effects altering both of the passive and active targeting processes.18–24 However, proper tuning of the NPs’ represents a chance for the optimization of the PC composition for enhancing the delivery of the NPs to the target tissue and provide an avenue for endogenous targeting.25,26
In the first part of this study,27 NPs possessing different physicochemical properties were synthesized. Polylactic co-glycolic acid (PLGA) NPs (NP1 and NP2) and lipid-polymer hybrid NPs (NP3 and NP4) were synthesized to constitute the uNPs. Both types of the NPs were then conjugated to the cRGDyk peptide at polymer to cRGDyk peptide (P:R) molar ratio of 1:1 to constitute the cNPs. cRGDyk peptide is a known targeting ligand to the αVβ3 integrin receptors overly expressed in cancer cells.28–30 Their PC have been characterized in terms of the total quantity and the relative abundance of distinct proteins of interest. This was followed by the characterization of the role of the PC on the NPs’ interaction with B16F10 melanoma cells in vitro under specific conditions (3 h incubation). VN protein adsorption from mouse serum (MS) has shown a potential ability to modulate higher NPs’ accumulation in the target cells which overexpress the VN receptor or the so-called αVβ3 integrin receptors.23,24 However, the experimental setting does not take into account the possibility of NPs’ elimination after their internalization or the occurrence of the process of exocytosis. Exocytosis and endocytosis have been reported to occur simultaneously; in which the internalized NPs can be excreted from the cells and re-internalized back in a continuous loop.31–33 In addition, the evaluation of the NPs’ behavior can generate false results of the intracellular accumulation when tested using an in vitro cell culture model. In this setting, the NPs are directly introduced to the cells, so the interaction between the two is virtually guaranteed. This model lacks the complex transport processes that the NPs face when they are applied in vivo, eg the distribution throughout the body, in normal and neoplastic tissues, the elimination by the RES organs and the transport through the highly complex tumor-associated extracellular matrix (ECM).
Therefore, in order to build stronger conclusions on the potential role of selected PC components in directing the NPs to the desired targets which can represent an appealing drug targeting approach, this part of the study investigates the in vitro uptake and elimination kinetics in B16F10 melanoma cells and the in vivo biodistribution and pharmacokinetics (PKs) in a melanoma mouse model.
Materials and Methods
Materials
Polylactic-coglycolic acid (PLGA) was purchased from Sigma Aldrich, Germany. Soybean lecithin was purchased from Roth, Germany. Fluorescein isothiocyanate (FITC) was also purchased from Santa Cruz, Germany. c(RGDyk) peptide 99.59% was purchased from Selleckchem, USA. Mouse melanoma cell line (B16.F10) was purchased from the American Type culture collection (ATCC; CRL-6475).
Female C57 Black 6 (C57BL/6) mice were obtained from the animal house breeds of the German University in Cairo (GUC). The experiment protocol followed ARRIVE guidelines and was approved by the Research Ethics Committee of the Faculty of Pharmacy and Biotechnology, GUC. Each three mice weighing 20–25 g were housed in suitably sized cages to allow free movements. The cages were exposed to standard conditions of housing with a 12-h light/dark cycle and a temperature of 22 °C. All mice received a standard laboratory diet and were granted free access to water. The mice were cared for on a daily basis. Mouse serum (MS) was obtained by collecting blood using the retro-orbital bleeding technique under aseptic conditions in specialized serum separator collection tubes. Animals were sacrificed by cervical dislocation and remains were disposed of by incineration according to the approved animal waste disposal system.
Evaluation of the Kinetics of the Intracellular Uptake and Elimination of the uNPs and cNPs
The concentration of intracellular NPs was determined in serum-free (SF) and serum-rich (SR) phenol red-free DMEM at predefined incubation times (30 min, 3, 6, 8 and 24 h) according to the protocol elaborated in part I27. In brief, B16F10 melanoma cells were seeded and incubated overnight under the standard conditions (37 °C and 5% CO2). A non-cytotoxic concentration of the fluorescently labelled NPs (250 µg/mL) was added under both SF and SR conditions. After the predefined incubation times, the concentration of the internalized NPs was calculated according to the fluorescence intensity of the labelling dye (Fluorescein isothiocyanate, FITC).
For the determination of the NPs’ uptake and elimination parameters, the experimental results were fitted in the one compartment extravascular-like model34–36 in Microsoft Excel 2016 and the corresponding pharmacokinetics add-in “PK Solver”37 according to the following equation; 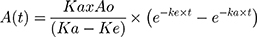 (Equation 1)
(Equation 1)
where A is the concentration of the internalized NPs; t is the time in hours; ka is the uptake rate constant in h−1; and ke is the elimination rate constant in h−1.
The area under the curve from 0 to 24 h post intravenous (IV) administration, AUC024 was calculated by the trapezoidal rule; AUC024 = 0.5 x (A0+A24) x (24–0) (Equation 2)
Competitive uptake of cNPs was performed by adding free cRGDyk (0.5 µg/mL) in both SF and SR culture media.
Evaluation of the in vitro Cellular Uptake of uNPs and cNPs by Fluorescence Cell Imaging
A volume of 300 µL of cells were seeded onto round glass coverslips (12 mm diameter) in 24-well plates at a density of 105 cells per well and incubated overnight at 37°C and 5% CO2. A suspension of fluorescein isothiocyanate (FITC)-loaded NPs in SF and SR phenol red-free DMEM achieving a final NPs’ concentration of 250 µg/mL was added after aspiration of the old medium and washing the cells with phosphate-buffered saline (PBS) to remove any debris. After the predefined incubation times (3 and 24 h), the medium was aspirated, cells were washed twice with cold PBS, and the cell membrane was stained with wheat germ agglutinin-AlexaFluor 594 (WGA-AF594) (Invitrogen, Germany) according to the manufacturer’s instructions. Cells were then washed with cold PBS twice again, fixed with 4% paraformaldehyde (PFA) (Elektron Technology, UK) and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI blue) (Sigma-Aldrich, Germany). The coverslips were then lifted, dried and mounted on glass slides using Mowiol mounting medium (Sigma-Aldrich, Germany) for the preservation of the fluorescence intensity and imaged using a Zeiss Axio star Plus fluorescence microscope (Carl Zeiss, Sweden).38
Evaluation of the in vivo Biodistribution of uNPs and cNPs
The biodistribution study was conducted according to the protocol designed by Yin et al as follows:39 mice were randomly grouped (n=3) and intradermally (ID) transplanted with B16F10 cells at a cell density of 0.5*106 cells in 50 µL of growth medium. The external tumor volume was monitored 2–3 times per week until it reached a volume of 100–130 mm3. The tumor volume was calculated as shown in equation (3).40
After reaching the target volume, mice were intravenously (IV) injected with 0.5 mL of the NPs suspended in PBS (10 mg/mL). After the predefined time points, mice were sacrificed by cervical dislocation under light anaesthesia and the required organs and tissues (heart, kidneys, liver, spleen, lungs and tumor) were collected and washed with a saline solution. Blood samples (0.5 mL) were withdrawn by cardiac puncture and placed in heparinized vials. Plasma was separated from the blood cells by centrifugation at 1500 x g for 5 minutes at 4 °C. All organs were then stored in Eppendorf tubes at −80 °C until further use. Organs were then thawed, weighed and homogenized in freshly prepared one molar (M) sodium hydroxide (NaOH) in a manual tissue homogenizer and diluted ten times then analysed for FITC at excitation/emission 494/517 nm using a spectrofluorophotometer RF 6000 (Shimadzu, Japan). Finally, calibration curves were constructed for each of the NPs for the quantitation of the NPs’ accumulation in different organs. A group of three mice were treated in a similar manner by injecting 0.5 mL saline to be used as controls.
The fluorescence of the tissue homogenates obtained from the treated or the control animals was normalized against the organ’s or tissue’s weight before subtraction of the auto-fluorescence for a fairer comparison of results. Finally, the biodistribution was expressed as the percentage of the injected dose per gram of the organ or tissue (% ID/g) at each time point.
Evaluation of the Pharmacokinetics (PKs) of uNPs and cNPs
PKs of the selected formulations were analysed using PK solver, an excel add-in37 to get a clearer insight on the influence of the NPs’ physicochemical properties and the cRDGyk peptide conjugation on their fate in the tumor-bearing mice and provide a rational approach for predicting the time course of the NPs in the vasculature and other tissues or organs.
The PK parameters studied in this work include the concentration at time zero (Co), the elimination half-life (t½E), the accumulation half-life (t½A), the volume of distribution (Vd), the clearance (Cl), the area under the curve (AUC) and the mean residence time (MRT).41
Statistical Analysis
The whole experiments were conducted at least in triplicate and the data is represented as the mean ± the standard error of the mean (SEM). The statistical analysis of data was performed using One-way ANOVA, Two-way ANOVA Tukey’s Multiple Comparison Test or t-test whenever appropriate at a confidence level of 95% using the GraphPad Prism 7.0 (GraphPad Software, Inc, San Diego, CA, USA). Levels of statistical significance are demonstrated as Asterisks (*).
Results
The Kinetics of the Intracellular Uptake and Elimination of uNPs and cNPs
In SF medium, all uNPs and cNPs showed a time-dependent accumulation inside the cells (Table 1 and Figure 1A and B). Only the polymeric uNPs, uNP1 and uNP2, (Table S1) showed a slight decrease in their intracellular concentration at 24 h. Upon the calculation of AUC024 by the trapezoidal method to indicate the overall accumulation of the NPs, similar levels of the uNPs were observed. However, uNP1 accumulated to a lower extent (P<0.5). On the other hand, the accumulation of the cNPs was comparably higher than their corresponding uNPs. The AUC024 of the polymeric cNP1 and cNP2 was more than 1.5-fold higher than that of the uNP1 and uNP2, respectively. Moreover, the hybrid cNP3 showed the highest increase in the intracellular accumulation recording a 2.5-fold higher AUC024 compared to the uNP3. In the meantime, the hybrid cNP4 showed the lowest increase under SF conditions recording a 1.2-fold higher accumulation than the uNP4.
 |
Table 1 The Parameters of the Uptake and Elimination Kinetics of uNPs and cNPs in SF and SR Conditions |
 |
Figure 1 The intracellular accumulation profiles of the uNPs (A and C) and the cNPs (B and D) under SF and SR conditions respectively. |
The NPs behaved differently in the SR media; a great decrease in the intracellular levels of the NPs was observed after 3 hours of incubation with the cells (Table 1 and Figure 1C and D). Therefore, the extravascular-like one-compartment model was used for the calculation of the uptake-related (ka) and the elimination-related (ke) parameters for a better understanding of the NPs’ endocytosis and exocytosis behavior, respectively. In addition, the Cmax and the AUC were used to indicate the overall accumulation of the NPs.
The polymeric formulation, uNP2, which had the smallest particle size in serum, showed the fastest rate of uptake. However, in cNPs, size was not a determining factor for the ka (all cNPs exhibited almost similar particle size in serum shown in Part I27 Table 2, available in Table S2 in supplemental materials). The NPs’ composition was observed to exhibit a more important role; the hybrid cNP4 showed a faster uptake rate than any of the other cNPs.
 |
Table 2 The Sum of the % ID/g of the Vascular Compartment (Plasma and Heart), the RES Organs (Kidneys, Liver and Spleen) and Lungs, and the Tumor Tissue for the uNPs and cNPs |
Regarding the effect of the peptide conjugation on ka, no significant difference in the rate of the intracellular uptake was observed for the cNP1 or the cNP3 in comparison to their corresponding uNPs. On the other hand,; there is a very significant (P<0.0001) reduction (32-fold) in the ka upon peptide conjugation in the polymeric cNP2 in comparison to uNP2. This effect accompanied the observed increase in the particle size in serum from 150 nm to 230 nm. On the contrary, the hybrid cNP4 showed a 13-foldincrease (P<0.0001) accompanying a similar increase in the particle size from 175 nm to 220 nm in serum.
Regarding the elimination rate constant (ke) in the polymeric NPs, it was observed that the large uNP1 showed a two-fold faster elimination rate than the smaller uNP2 (P<0.01). This correlates with the two-fold higher cumulative release of FITC observed for uNP1, as shown in Part I.27 An opposite trend was observed for the hybrid NPs; the smaller uNP4 showed almost a 5-fold higher elimination rate than uNP3 (P<0.01). This opposes the release results in which the uNP3 recorded a higher cumulative release of FITC than the uNP4. This could indicate that the NPs were exocytosed in their intact form. On the other hand, similar elimination patterns were recorded for all the cNPs. Upon the comparison of the uNPs with cNPs, it could be concluded that the peptide conjugation caused a reduction in the elimination rate as could be observed by the 16-, 13-, 2- and 26-fold decrease in the ke values for the cNP1, cNP2, cNP3 and cNP4, respectively.
It is worth highlighting that the decrease in the fluorescence intensity in the intracellular uptake study could be attributed to the elimination of the intact FITC-loaded NPs or the free dye released from the NPs after their internalization. Therefore, the kinetics of the uptake and elimination of the free FITC dye has been evaluated and discussed under SF and SR conditions in the supplementary materials results and discussion section 1.2 (Figure S1 a & b, respectively, and Table S3). The results were then compared with the in vitro release results of the NPs’-loaded FITC (shown in Part I27).
The maximum concentration (Cmax) of the uNPs and the cNPs followed the following order; uNP4> uNP3> uNP1 > uNP2 and cNP3> cNP4> cNP1 ≈ cNP2, respectively. The hybrid NPs, NP3 and NP4, in their both forms, unconjugated and peptide-conjugated, showed higher levels of accumulation in the cells than the polymeric NPs, NP1 and NP2. Moreover, the peptide conjugation affected the accumulation of the NPs significantly recording a 2–4-fold higher Cmax in cNPs compared to their corresponding uNPs.
The area under the curve (AUC) of the hybrid NPs (NP3 and NP4) indicated a significantly higher accumulation (P<0.001) than the polymeric NPs (NP1 and NP2) in both of their conjugated and unconjugated forms.
Collectively, the effect of serum proteins on the NPs’ uptake was more evident in the uNPs. uNPs showed a much higher accumulation in SF conditions (P<0.0001) than SR conditions. The polymeric NPs (uNP1 and uNP2) and the hybrid (uNP3 and uNP4) showed a 7- and 3-fold higher accumulation in SF over SR conditions, respectively. On the contrary, the cNPs were generally less affected by the presence of serum proteins. The extent of the accumulation of the NPs was even reversed in the case of cNP4; it recorded a slightly higher accumulation in the SR medium.
The competitive uptake was evaluated for the cNPs using free cRGDyk peptide (0.5 µg/mL) for the saturation of the integrin receptors in both SF and SR media. Results showed more than 50% reduction in the Cmax in all the NPs indicative of the dependence of the uptake of the cNPs on the integrin receptor-mediated endocytosis (data not shown). Moreover, cNPs were also prepared at P:R molar ratio of 1:10 and 1:50 in order to evaluate the effect of the peptide conjugation density on the NPs’ behaviour. The physicochemical properties of the cNPs prepared at different conjugation densities and the parameters of the intracellular uptake and elimination kinetics are shown in the supplemental materials results and discussion section 1.1 (Table S1), and section 1.2 (Table S4 & Figure S2 a-d) respectively.
Fluorescence Cell Imaging
The intracellular uptake of the NPs under both SF and SR conditions was evaluated by fluorescence cell imaging. The intensity of the green fluorescence of FITC observed in supplementary materials results and discussion section 1.3 (Figures S3–6) matched the obtained results of the quantitative uptake (Table 1 and Figure 1). The extent of the NPs’ accumulation after 24 h incubation was higher than 3 h in SF conditions and lower in SR conditions.
Biodistribution and Pharmacokinetics (PKs) of uNPs and cNPs in B16F10 Melanoma Mouse Model
Comparison of the Biodistribution Profiles of uNPs and cNPs in Selected Organs
Different NPs formulations, uNPs and cNPs, showed different patterns of biodistribution in the selected organs as shown in Figure 2. However, for an easier interpretation of the data, the % ID/g results obtained from the heart and the plasma were grouped to represent a vascular compartment indicating the circulation time of the NPs or their affinity to the microtubular structures of the heart muscles.42 Similarly, the results of the RES organs (kidney, liver and spleen)43 were grouped with the results of the lungs which were reported to accumulate NPs after being sequestered by lung macrophages in a similar manner to RES uptake.42,44 Finally, the results of the melanoma tumor tissue represented the target tissue affected by the EPR effect and the active targeting upon peptide conjugation of NPs45 (Table 2).
The % ID/g results showed that the polymeric uNPs, uNP1 and uNP2 (Figure 2A–C, D–F respectively), were rapidly cleared from the vascular compartment after 24 h post the IV injection. On the contrary, the hybrid uNPs, uNP3 and uNP4 (Figure 2G–I, J–L respectively), maintained relatively higher % ID/g for longer period of time. Additionally, both uNP1 and uNP2 showed higher % ID/g in the kidneys after 2 h than uNP3 and uNP4, whereas uNP1 possessed the highest interaction with the RES organs followed by uNP2. On the other hand, uNP3 and uNP4 showed a minimal accumulation in the RES organs and lungs recording less than 15% ID/g after 6 h.
The tumor uptake of the NPs was evaluated over time and showed the following patterns. After 2 h post-administration, small amounts of the NPs were detectable followed by a steep increase at 6 h and then a decline of up to 24 h. uNP4 exhibited the highest localization in the tumor tissue followed by uNP1 then uNP2, while uNP3 showed the least accumulation in the tumor over the tested time points.
The peptide conjugation for the active targeting of melanoma altered the biodistribution patterns for the grouped compartments shown in Table 2. It generally reduced % ID/g in the organs of the RES and the lungs. Additionally, the peptide conjugation caused a reduction in the % ID/g in the vascular compartment in the case of NP1 and NP2 (Figure 2A–C, D–F respectively). However, cNP4 showed an increased accumulation in the vascular compartment at all the tested time points.
Regarding the effect of the peptide conjugation on the tumor accumulation, all the cNPs showed a higher accumulation than the corresponding uNPs. The most pronounced increase after 2 h was recorded for cNP1 and cNP4 (Figure 2A–C, J–L respectively) which showed more than a 10-fold higher % ID/g than the corresponding uNPs. On the other hand, after 6 h, cNP1 showed a 5-fold decrease in the NPs’ accumulation. In contrast, cNP2 and cNP4 continued to show an enhancement in the tumor accumulation that extended to 24 h, while no marked enhancement in the NPs’ accumulation in the tumor was observed in the case of cNP3 (Figure 2G–I).
Pharmacokinetics (PKs)
The one compartment model was selected for the calculation of the PK parameters of all the uNPs and cNPs in the supplementary materials results and discussion section 1.4 (Figure S7 a-e, g-h) except cNP3 which continued to show an increased % ID/g in the plasma over time with no elimination phase (Figure S7-f); therefore, the linear trapezoidal method was utilized to calculate the AUC024.
Upon the comparison of the PKs behaviour of the uNPs, the smaller NPs formulation, uNP2 (~150 nm) showed the longest t1/2E and MRT than both of uNP1 and uNP3 which possessed bigger particle size (~200 nm) (Part I27-Table 2, available in Table S2 in supplemental material). uNP2 also showed the largest Vd and the lowest AUC than both of uNP1 and uNP3 which were statistically indifferent (Table 3).
 |
Table 3 The PK Parameters of the uNPs and the cNPs in the Vascular Compartment (the Plasma and the Heart) |
The peptide conjugation caused a delayed elimination as witnessed by the larger t1/2E in the polymeric cNPs, cNP1 and cNP2, and the absence of an elimination phase in the hybrid cNP3. In the meanwhile, the peptide conjugation caused an increased accumulation of the NPs in the plasma, most pronounced in the case of the hybrid cNP3 and cNP4, as observed from AUC024. However, cNP1 showed a lower AUC and a higher Vd.
uNP3 showed a continuous increase in the % ID/g with no detectable elimination phase (Figure S8e); therefore, the linear trapezoidal method was utilized to calculate the AUC024. Results showed no significant difference (P>0.05) in the % ID/g of uNP1, uNP2 and uNP4 (Table 3 and Figure S8 a,c & g respectively). In the meanwhile, the polymeric formulations; uNP1 and uNP2 (Figure S8 a,c respectively), showed a faster accumulation in the heart as shown by the lower t1/2A and a faster elimination as shown by the lower t1/2E (P<0.05) which eventually resulted in a lower MRT than the hybrid ones, uNP3 and uNP4.
Upon the peptide conjugation, all cNPs (Figure S8 b,d,f & h) showed faster accumulation and elimination rates than their corresponding uNPs. In addition, the cNPs recorded lower levels in the heart (AUC) and shorter MRT.
uNP3 showed a continuous increase in the % ID/g (Figure S9 e); however, the AUC024 of the hybrid NPs, uNP3 and uNP4 (Figure S9 e & g respectively) was significantly lower than the polymeric ones, uNP1 and uNP2 (P<0.001 and P<0.05, respectively) with almost a 2.5- and a 1.5-fold decrease as shown in (Table 4 and Figure S9 a & c respectively). Regarding the polymeric uNPs, the small uNP2 showed a lower accumulation as evidenced by Cmax and AUC (P<0.01) than the larger uNP1.
 |
Table 4 The PK Parameters of the uNPs and the cNPs in the RES (the Kidney, the Liver and the Spleen) and the Lungs |
Upon peptide conjugation, all the cNPs (Figure S9 b,d,f & h) showed faster accumulation (small t1/2A); however, lower levels of NPs were recorded (smaller Cmax and AUC) when compared to their unconjugated counterparts.
The hybrid uNPs, uNP3 and uNP4 (Figure S10 e & g respectively) showed significantly lower levels (P<0.01) in the liver than the polymeric uNPs, uNP1 and uNP2 (lower Cmax and AUC) (Table 4 and Figure S10 a & c, respectively).
The levels of NPs were reduced upon the peptide conjugation in all the formulations (Figure S10 b, d,f &h).
The small uNP2 and the hybrid uNP3 and uNP4 (Table 4 and Figure S11 c, e & g respectively) recorded lower Cmax and AUC in the spleen than the large polymeric uNP1 (Table 4 and Figure S11 a).
The peptide conjugation caused a reduction in NPs’ levels in all the formulations except cNP2, which showed a continuous increase with no elimination phase (Table 4 and Figure S11 b, d,f & h).
The hybrid uNPs, uNP3 and uNP4 (Table 4 and Figure S12 e & g respectively), showed significantly lower Cmax and AUC (P<0.05) in the lungs than the polymeric uNPs, uNP1 and uNP2 (Table 4 and Figure S12 a & c, respectively).
The levels of the NPs were reduced upon the peptide conjugation in all formulations (Table 4 and Figure S12 b,d,f & h).
uNP4 (Figure S13 g) showed the highest levels in the tumor tissue as denoted by the Cmax followed by uNP1 and uNP2 (Figure S13 a & c, respectively), while uNP3 (Figure S13 e) recorded the lowest Cmax and the slowest accumulation rate (t1/2A).
The peptide conjugation of cNP1 caused a faster accumulation in the tumor tissue to achieve almost a 1.7-fold higher Cmax at a much shorter tmax (0.4 h); however, the elimination rate upon the peptide conjugation increased causing an overall decrease in the NPs’ accumulation (AUC) and MRT (Table 5 and Figure S13 b).
 |
Table 5 The PK Parameters of the uNPs and the cNPs in the Tumor Tissue |
In the smaller polymeric cNP2, the peptide conjugation slightly reduced the accumulation rate (longer t1/2A); however, the NPs’ accumulation extent (as denoted by the Cmax and the AUC) was more than a 3.5-fold higher than the corresponding uNP2. On the other hand, the elimination was slightly reduced causing a slight increase in the MRT (Table 5 and Figure S13 d).
The peptide conjugation of the hybrid cNP3 (Table 5 and Figure S13 f) caused an insignificant increase (P>0.05) in the rate and the extent of NPs’ accumulation in the tumor tissue, while the hybrid cNP4 showed a much higher extent of the accumulation in the tumor tissue than all the other formulations (Table 5 and Figure S13 h).
The Effect of the Selected Serum Proteins in the PC of uNPs and cNPs on the Biodistribution and the PKs
As previously illustrated from the NPs’ PC analysis (Part I27), VN levels were generally higher for the cNPs than the uNPs. This finding goes in harmony with the observed accumulation of the NPs in the tumor cells as shown in the in vitro cellular uptake (Table 1) and the tumor tissue as shown in the in vivo biodistribution and PKs (Tables 2 and 5). For example, it was observed that NP4 exhibited high levels of VN in both of its conjugated and unconjugated forms which could justify its intensified accumulation in the tumor tissue on both of the in vitro and in vivo levels. On the contrary, NP3, in both of its conjugated and unconjugated forms, exhibited the lowest levels of VN in the adsorbed corona, which could also correlate with the tumor accumulation tendencies.
In the case of uNPs, uNP1 and uNP4 showed a similar low abundance of C3 in their PCs; however, they showed a different accumulation in the RES organs. The NPs’ accumulation was the highest in the case of uNP1 and lowest in the case of uNP4. Upon the peptide conjugation, cNP2 showed the highest abundance of C3 that correlated with the highest RES organs’ accumulation relative to the other cNPs (Tables 2 and 4).
The low C3 abundance in uNP1 and uNP4 correlated with a high AUC024 in plasma, while in cNPs, the hybrid lipid-polymer formulations, cNP3 and cNP4, which showed low abundance of C3 showed a high accumulation in plasma (AUC024) in Table 3.
Low abundance of CLU was shown in the polymeric uNPs, uNP1 and uNP2, which correlate inversely with their accumulation in the RES organs. They showed a total % ID/g of 39.8% and 28.7% respectively after 6 h (Table 2). Upon the peptide conjugation, CLU adsorption markedly increased on the polymeric cNPs, cNP1 and cNP2, and the RES organ accumulation of the same NPs decreased significantly. Regarding the hybrid uNPs, uNP3 and uNP4, a high abundance of CLU was observed coinciding with their low accumulation in the RES organs. However, upon the peptide conjugation, CLU levels decreased while their accumulation in the RES organs continued to decrease as observed from the AUC024 (Tables 2 and 4).
The peptide conjugation, which caused CLU to deposit to a greater extent on the polymeric cNPs and to a lesser extent on the hybrid ones in comparison to their corresponding uNPs, caused a delayed elimination from the vascular compartment as witnessed by a longer t1/2E (cNP1, cNP2 and cNP4) or an absence of the elimination phase (cNP3). Additionally, an increased accumulation of the NPs in the plasma was observed for the hybrid cNP3 and cNP4 (higher AUC024) (Table 3).
Discussion
For the intracellular uptake studies in B16F10 melanoma cells in vitro in SF conditions, the linear trapezoidal method was utilized for the calculation of the AUC024. AUC024 indicates the NPs’ accumulation in the cells over the tested duration (Table 1 and Figure 1). A time-dependency of the uptake has been observed in SF conditions consistently with the previous reports that have shown the time- and concentration-dependency of PLGA NPs’ uptake via endocytosis.46 In the absence of serum, all the uNPs showed similar accumulation in the melanoma cells except for uNP1. It showed a slight decline at 24 h attributable to the activation of the exocytosis process.47 Upon the peptide conjugation, all the cNPs showed higher accumulation in the cells, most prominently in the hybrid cNP3. This could be attributed to the role of both of the cRDGyk peptide and the lipid coat in increasing the cellular levels of the NPs.48,49 The competitive inhibition of the cNPs’ uptake when the free cRGDyk peptide was utilized proves the involvement of the integrin receptor in the endocytosis process of the NPs. This is consistent with the previous reports showing a competitive inhibition of NPs' uptake in the presence of RGD peptide in a concentration-dependent manner.50
In SR conditions, the one-compartment PK analysis was performed using PK solver add-in of Microsoft Excel 2016 in order to have a more accurate comparison of the uptake and the elimination parameters of uNPs and cNPs. The model selection has been justified in the view of the previous studies. The IV infusion-like calculations were applied for the calculation of the PK parameters for NPs whose constant flux was ceased and their elimination was observed over time.34,35 The monod-like calculations have been utilized in another study when a saturation–like performance of the NPs’ uptake was observed.36 In our study, the extravascular-like calculations of the uptake and the elimination kinetics were performed for the NPs tested in SR conditions, whereas the NPs’ exposure to the cells was not ceased and no saturation or steady-state accumulation of the NPs was observed. What made us think that this model is more explanatory to our results is that the NPs’ uptake and elimination are in harmony; the internalized NPs could exit the cell via different pathways33 and be re-internalized in a continuous loop.31,32 The time-dependency of the uptake of the NPs was broken in the presence of serum; there was a decrease in the level of the NPs in melanoma cells after 3 h. Therefore, the accumulation of NPs was found to depend on a balance between the uptake and the elimination processes. It could also be suggested that the receptor recycling and the exocytosis play a role as early as 3 h or less after the incubation of the NPs with the cells as previously reported.47,51,52 In a study by Cui et al, PEGylated gold NPs were rapidly exocytosed, especially when conjugated to a cRGD-based peptide, under SR conditions. This was correlated with the rapid recycling of the αvβ3 integrin receptor.47 In another study by Tedja et al, the presence of serum was found to regulate the continuity of the endocytosis and exocytosis loop of the titanium dioxide (TiO2) NPs. In the SR conditions, TiO2 NPs were rapidly uptaken in the first two hours after incubation with the lung cancer cells, then significantly exocytosed in the next four hours. A second wave of uptake was observed resulting in an increased NPs’ accumulation in the cells.53 In addition, Reinholz et al have identified a putative proteome profile necessary for regulating the exocytosis of the NPs from Caco-2 cells.54
The difference in the uptake and the elimination parameters of the different NPs could be attributed to the dependency of both the endocytosis and the exocytosis on different parameters including the particle size,33,55,56 the composition or the rigidity of the NPs,57 and the mechanism of the intracellular uptake;46,58,59
Some studies have attributed the higher uptake of the small-sized NPs, eg uNP2 in our study, to their larger surface area and the higher contact with the biological membranes.33,36,60–62 For example, Guarnieri et al have reported that the 49 nm-sized NPs showed 4-fold higher uptake than 100 nm-sized NPs.36 This advantage of the uNP2 was lost upon the peptide conjugation, whereas the cNP2 increased in size and showed a 32-fold decrease in its ka. On the other hand, cNP4, which showed a similar increase in the particle size, showed an opposite pattern in ka. This could be explained by the difference in the composition of the NPs that could also be a determining factor of the cellular uptake. The incorporation of a lipid coat onto PLGA nanoparticles was reported to increase the cellular internalization of the NPs and their targeting abilities.49 For example, Guo et al reported higher uptake of the lipid-coated PLGA NPs over the uncoated ones in A549 lung cancer cells.63 In addition, the mechanism of the intracellular uptake has proven to contribute to a great extent to both uptake and the elimination of the NPs. In part I of this study,27 the evaluation of the endocytic mechanism of the NPs’ uptake has been performed. The results suggest that the NPs are internalized via an energy-dependent pathway. In the case of the hybrid uNPs, uNP3 and uNP4, a cholesterol-dependent uptake mechanism dominated especially under SR conditions. While in the case of the polymeric uNPs, uNP1 and uNP2, a caveolin-mediated uptake mechanism was more prominent. Moreover, the peptide conjugation was shown to affect the mechanism of NPs’ uptake and increase the reliance on caveolin rather than clathrin-dependent pathway. These observations are consistent with previous studies showing the dependence of the mechanism of the NPs’ uptake on their physicochemical properties.64,65 PC formation could as well justify the controversies in the difference in the uptake and the elimination kinetics of the uNPs and the cNPs in the SF and SR conditions. The lower extent of the accumulation of the uNPs (lower AUC), especially the polymeric ones (uNP1 and uNP2) and to a lesser extent the hybrid ones (uNP3 and uNP4) in the cells in the presence of serum proteins could be explained in different ways. First, the endocytosis of the NPs is less efficient in the presence of serum, while the exocytosis is more pronounced.66 The PC could also decrease the NPs’ adhesion onto the cell membrane.23,25 On the contrary, the high extent of accumulation of the cNPs in the cells in the presence of serum proteins was also observed and was consistent with the previous reports that showed the cRGD peptides’ ability to maintain their active targeting efficacy in the presence of the PC . This effect was more pronounced in the large-sized NPs which possessed multiple binding sites.25,67,68 In addition, some proteins in the PC could play a role in regulating the NPs’ uptake or elimination.25,69–71 For example, the cNP4, which showed the highest abundance of VN in their PC, showed the highest accumulation in the cells. This is consistent with previous reports that elaborated the role of VN in regulating the uptake in integrin receptor overexpressing cells. In a study by Ritz et al, a positive correlation between the enrichment of VN in the PC and the intracellular uptake was observed.70 In addition, VN has been identified as an important PC fingerprint correlating with the NPs’ cell adhesion by Palchetti et al.72 Furthermore, DOTAP cationic liposomes tended to enrich VN that facilitated their uptake in cancer cells expressing high levels of VN ανβ3 integrin receptor.73 VN adsorption did not only affect the NPs’ internalization but was also reported to affect the downstream biological effects.74
Similarly, the abundance of clathrin heavy chain protein (CHC) in the PC of the hybrid cNPs, cNP3 and cNP4, could have contributed to the higher accumulation of these formulations by a clathrin-mediated endocytosis.12,23,61 On the contrary, annexin I (ANXA1), which has also been exclusively found in the PC of the hybrid cNPs, cNP3 and cNP4, was reported to be associated with the receptor recycling and exocytosis.66 However, these two formulations showed low elimination rates owing to the proton sponge effect that facilitates the endolysosomal escape which could possibly override the role of ANXA1 in the PC. This effect is imparted by the polyamines which can neutralize the endosomes, cause disruption of their membranes and mediate the endolysosomal escape.12 This effect could justify the decrease in the ke observed for the cNPs when compared to the corresponding uNPs.
In the biodistribution study performed for the eight different formulations of the NPs, it could be observed that results of the % ID/g do not add up to 100% (Table 3) indicating the involvement of other tissues not assayed in this work, eg lymphatic system, genital system, gastrointestinal tract (GIT), etc., or the adhesion of the NPs to the endothelial walls of the blood vessels which could constitute up to 50% of the injected dose.45,75 All the organs showed a gradual decline in the % ID/g over time. This could be attributed to the elimination or the redistribution of the NPs to other organs or tissues.76 The higher levels of the NPs detectable in the vascular compartment in the hybrid uNPs, uNP3 and uNP4, could indicate the role of the lipid coat in prolonging the blood circulation time of the NPs and reducing their RES uptake.49 This finding is similar to the plasma circulation time data obtained for PLGA and lipid-PLGA hybrid NPs reported by Godara et al.77 Upon the evaluation of the PKs, the small uNP2 possessed a fast equilibration in the plasma with no absorption-like phase, a larger Vd, and a smaller AUC. Similar findings were reported by Rafiei et al in which the small-sized PLGA NPs (120 nm) demonstrated higher serum concentrations early after the IV administration, while the relatively large-sized PLGA-PEG NPs (190 nm) exhibited a higher concentration afterwards owing to the hydrophilicity imparted by the PEG.42 uNP2 also showed a slower elimination rate than the other uNPs; this could be attributed to its smaller size as previously reported.78 On the contrary, uNP1, uNP3 and uNP4 showed a delayed peak in the plasma which could be attributed to the adhesion of the NPs to the blood vessels’ walls at the site of injection.45,75 A similar delay in the peak was observed for chitosan-coated gold NPs.79 The peptide conjugation caused a delayed elimination as witnessed by a larger t1/2E in cNP1, cNP2 and cNP4 and an absence of the elimination phase in cNP3. A similar observation was previously reported in which the EGFR-targeted NPs prolonged the plasma and the tumor half-lives and the MRT of lonidamine relative to the non-targeted NPs.15
The higher NPs’ accumulation detectable at 6 h for the polymeric uNPs, uNP1 and uNP2, in the kidneys (Figure 2B and E) and the higher AUC (Table 4 and Figure S9) could be attributed to the faster degradation of the polymeric NPs. It has been shown that the presence of lipids in the composition of the PLGA NPs could retard the degradation of the PLGA chains in vivo.80 The faster degradation could promote a faster elimination of the released dose of the drug from the NPs.75 In a study by Navarro et al, 100–140 nm-sized PLGA and PLGA/chitosan NPs showed the second highest accumulation in the kidneys. This was attributed to the possibility of the accumulation of fragments of the fluorescently conjugated NPs in the kidneys on account of the polymer’s degradation.81
The high interaction of the uNP1 with the RES organs and the lungs which was more than uNP2 and the hybrid uNPs, uNP3 and uNP4, could be explained by the effect of smaller particle size in the case of uNP2 and the presence of a biocompatible lipid coat in the hybrid uNP3 and uNP4. Smaller-sized NPs were previously reported to exhibit a longer circulation time and a lower RES uptake,13,42,75 while the lipid coat was reported to impart some deformability to the NPs in the case of uNP3 and uNP4 which decreases the interaction with the macrophages and hence reduces the RES uptake.44,49 However, the uNP1 was easily taken up by the macrophages of the RES because of their stiffness in comparison to uNP3 as previously reported.13,82 Therefore; similarly, sized NPs (uNP1 and uNP3) and (uNP2 and uNP4) showed different biodistribution patterns owing to the difference in their surface properties and deformability.
Regarding the tumor accumulation of the uNPs, uNP1 and uNP4 exhibited an advantage over the other two formulations; this could be explained by virtue of the relatively larger size of uNP1 over uNP2 which secures multiple ligand-to-receptor interactions that could bring about more efficient internalization. On the contrary, smaller-sized NPs required a critical threshold of NPs density on the cell membrane to trigger the internalization process.4,59 On the other hand, uNP4 was superior in the tumor accumulation over the other formulations owing to the advantage imparted by the biocompatible lipid coat. It was previously reported that the hybrid lipid-polymer NPs exhibit better cellular accumulation and targeting properties over the polymeric ones.49,63
It was found that the conjugation to the cRGDyk peptide reduced the levels of the NPs in the organs of the RES and the lungs probably because they increased in the tumor. The targeting ligands could trigger the internalization by melanoma cells after extravasation into the tumor tissue.16 Similar reduced levels of the NPs in the kidneys and the lungs were reported by Yu et al upon amine modification of mesoporous silica NPs.83 In addition, the lowered levels in all the organs assayed in this study after 6 h (lower total % ID/g) could raise a question of the involvement of other un-assessed organ(s) or tissue(s)45,75 probably due to alteration of NPs physicochemical properties (particle size and surface charge) upon the peptide conjugation (Part I27-Table 2- available in Table S2 in the supplemental material).
The ability of the cNPs (especially cNP1, cNP2 and cNP4) to maintain a significantly higher concentration in the tumor tissue could possibly be related to the uptake by the targeted melanoma cells which over-express αvβ3 integrin receptors. The uNPs, without the cRGDyk peptide, could possibly have diffused away from the tumor tissue over time in the absence of a sufficient cell uptake.43 On the other hand, the tumor accumulation was observed to be low in the case of the uNP3. It even only slightly increased upon the peptide conjugation. It is possible that NP3, in both of its unconjugated and peptide-conjugated forms, has a low diffusion across the tumor’s leaky vasculature. It has been shown in previous work that passive and active targeting are complementary and that passive targeting is a prerequisite for active targeting.3 In other words, the NPs have to reach the tumor site through the leaky vasculature in order to promote the interaction between the targeting ligands and the target site.4,16 Another possible explanation for the insignificant increase in the tumor accumulation of the NPs in the case of cNP3 could be the detachment of the peptide functionalized lipid coat from the PLGA core during the NPs’ distribution phase leading to the failure of active targeting to the tumor site as reported previously by Jack Hu et al.84 In addition, it has been reported for some NPs that the adsorption of PC can completely shield the chemically coupled targeting moieties.26
Regarding the effect of the relative abundance of the selected serum proteins in PC of the NPs on their in vivo behaviour, different roles could be concluded. As elaborated earlier in the discussion of the in vitro intracellular uptake, VN abundance in the PC is positively correlated with tumor accumulation in vivo as well. This is consistent with the discussed role of VN as a ligand for ανβ3 integrin receptor or the so-called VN-receptor that is overexpressed on melanoma cells.22,25,69,70,85
The positive correlation between C3 accumulation in the NPs’ adsorbed PC and the high interaction with the RES organs or the fast clearance from the vascular compartment could be explained in the light of the opsonization process. Specific proteins such as immunoglobulins and complement proteins are known to mediate the interactions with the phagocytic cells and influence the clearance process.22,85 This phenomenon has been utilized by Shen et al, to target the B cells in the spleen. It was found that the deposition of C3 on the dextran-coated ferrous NPs promoted their accumulation in the B cells in a complement-dependent manner.86 In another study, time evolution of the amount of C3 on NPs’ surfaces directly correlated with increased accumulation of the NPs in the liver mediated by Kupffer cells.87 On the other hand, opposite patterns were observed in the case uNP1 which showed a low abundance of C3 and a high RES organs’ accumulation. This could be explained on the basis of the presence of other types of proteins which could influence the interaction with the RES organs or affect the vascular clearance times.41,88
On the contrary to C3 which is an opsonin, CLU is categorized as a dysopsonin. It shields the NPs from being opsonized, reduces the RES uptake, and consequently increases the circulation time.22,41,68,88–90 The abundance of CLU in the PC of PLGA NPs, poly ethyl ethylene phosphate (PEEP)-coated polystyrene or silver NPs was previously reported to significantly reduce the clearance rates41 and the nonspecific uptake by the macrophages91 or the macrophage-like THP-1 cells, respectively.92 This justifies the low-RES organs’ accumulation and the longer circulation time for the NPs showing a high abundance of CLU and vice versa for those with low abundance of the protein in their PC.
Conclusion
The highly heterogeneous nature of the passive and the active targeting of cancer therapeutics contributed to the failure of achieving convincing results in the clinical trials. To this end, tremendous efforts have been exerted to understand the source of heterogeneity and led to the understanding that there is no one-size fits all in nanomedicine. This has recently paved the road for revolutionizing nanomedicine from passive targeting to personalized nanomedicine. PC deposition on the NPs after systemic administration has contributed to a great extent to the lack of the in vitro-in vivo correlation and the preclinical to clinical extrapolation.
In our study, NPs possessing variable physicochemical properties tended to recruit cellular and serum proteins to variable extents. The abundance of VN in the PC of some NPs has been correlated with a high extent of tumor accumulation in both the in vitro and the in vivo settings. In addition, the recruitment of an opsonin (C3) and a dysopsonin (CLU) to different degrees has been shown to alter the extent of the NPs interaction with the RES and the blood circulation time. Therefore, it could be hypothesized that the modulation of the PC via tuning of the NPs’ properties could serve as a new hope for achieving endogenous targeting of nanotherapeutics. In this aspect, our future outlook is the investigation of the potential of selective recruitment of VN to achieve cancer targeting and C3 protein to achieve RES targeting via ligand–target interaction to evaluate if this can possess an advantage over tuning the NPs’ physicochemical properties.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Dasgupta A, Biancacci I, Kiessling F, Lammers T. Imaging-assisted anticancer nanotherapy. Theranostics. 2020;10(3):956–967. doi:10.7150/thno.38288
2. Golombek SK, May J-N, Theek B, et al. Tumor targeting via EPR: strategies to enhance patient responses. Adv Drug Deliv Rev. 2018;130:17–38. doi:10.1016/j.addr.2018.07.007
3. Danhier F, Breton AL, Préat V. RGD-based strategies to target alpha(v) beta(3) integrin in cancer therapy and diagnosis. Mol Pharm. 2012;9(11):2961–2973. doi:10.1021/mp3002733
4. Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14(1):1–16. doi:10.1146/annurev-bioeng-071811-150124
5. Mundra V, Li W, Mahato RI. Nanoparticle-mediated drug delivery for treating melanoma. Nanomedicine (Lond). 2015;10(16):2613–2633. doi:10.2217/nnm.15.111
6. Kwon IK, Lee SC, Han B, Park K. Analysis on the current status of targeted drug delivery to tumors. J Control Release. 2012;164(2):108–114. doi:10.1016/j.jconrel.2012.07.010
7. Manzano M, Vallet-Regí M. Mesoporous silica nanoparticles for drug delivery. Adv Funct Mater. 2020;30(2):3–5. doi:10.1002/adfm.201902634
8. Prabha S, Arya G, Chandra R, Ahmed B, Nimesh S. Effect of size on biological properties of nanoparticles employed in gene delivery. Artif Cells Nanomed Biotechnol. 2016;44(1):83–91. doi:10.3109/21691401.2014.913054
9. Zhang Y, Liu AT, Cornejo YR, Van Haute D, Berlin JM, Tortiglione C. A systematic comparison of in vitro cell uptake and in vivo biodistribution for three classes of gold nanoparticles with saturated PEG coatings. PLoS One. 2020;15(7):e0234916. doi:10.1371/journal.pone.0234916
10. Liu H, Wang J, Li W, Hu J, Wang M, Kang Y. Cellular uptake behaviors of rigidity-tunable dendrimers. Pharmaceutics. 2018;10(3):99. doi:10.3390/pharmaceutics10030099
11. Sun J, Zhang L, Wang J, et al. Tunable rigidity of (Polymeric Core)-(Lipid Shell) nanoparticles for regulated cellular uptake. Adv Mater. 2015;27(8):1402–1407. doi:10.1002/adma.201404788
12. Yameen B, Choi WI, Vilos C, Swami A, Shi J, Farokhzad OC. Insight into nanoparticle cellular uptake and intracellular targeting. J Control Release. 2014;190:485–499. doi:10.1016/j.jconrel.2014.06.038
13. Xu J, Gattacceca F, Amiji M. Biodistribution and pharmacokinetics of EGFR-targeted thiolated gelatin nanoparticles following systemic administration in pancreatic tumor-bearing mice. Mol Pharm. 2013;10(5):2031–2044. doi:10.1021/mp400054e
14. Voigt J, Christensen J, Shastri VP. Differential uptake of nanoparticles by endothelial cells through polyelectrolytes with affinity for caveolae. Proc Natl Acad Sci. 2014;111(8):2942–2947. doi:10.1073/pnas.1322356111
15. Milane L, Duan Z, Amiji M. Biodistribution and pharmacokinetic analysis of combination lonidamine and paclitaxel delivery in an orthotopic animal model of multi-drug resistant breast cancer using EGFR- targeted polymeric nanoparticles. Nanomedicine. 2011;7(4):435–444. doi:10.1016/j.nano.2010.12.009
16. Chen WC, Zhang AX, Li S-D. Limitations and niches of the active targeting approach for nanoparticle drug delivery. Eur J Nanomed. 2012;4(2–4):89–93. doi:10.1515/ejnm-2012-0010
17. Francia V, Yang K, Deville S, Reker-Smit C, Nelissen I, Salvati A. Corona composition can affect the mechanisms cells use to internalize nanoparticles. ACS Nano. 2019;13(10):11107–11121. doi:10.1021/acsnano.9b03824
18. Gossmann R, Fahrländer E, Hummel M, Mulac D, Brockmeyer J, Langer K. Comparative examination of adsorption of serum proteins on HSA- and PLGA-based nanoparticles using SDS–PAGE and LC–MS. Eur J Pharm Biopharm. 2015;93:80–87. doi:10.1016/J.EJPB.2015.03.021
19. Weber C, Simon J, Mailänder V, Morsbach S, Landfester K. Preservation of the soft protein corona in distinct flow allows identification of weakly bound proteins. Acta Biomater. 2018;76:217–224. doi:10.1016/j.actbio.2018.05.057
20. Liu N, Tang M, Ding J. The interaction between nanoparticles-protein corona complex and cells and its toxic effect on cells. Chemosphere. 2020. doi:10.1016/j.chemosphere.2019.125624
21. Hu Z, Zhang H, Zhang Y, Wu R, Zou H. Nanoparticle size matters in the formation of plasma protein coronas on Fe3O4 nanoparticles. Colloids Surf B Biointerfaces. 2014;121:354–361. doi:10.1016/j.colsurfb.2014.06.016
22. Simon J, Müller LK, Kokkinopoulou M, et al. Exploiting the biomolecular corona: pre-coating of nanoparticles enables controlled cellular interactions. Nanoscale. 2018;10(22):10731–10739. doi:10.1039/C8NR03331E
23. Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6(7):5845–5857. doi:10.1021/nn300223w
24. Cagliani R, Gatto F, Bardi G. Protein adsorption: a feasible method for nanoparticle functionalization? Materials. 2019;12(12):1991. doi:10.3390/ma12121991
25. Chen D, Ganesh S, Wang W, Amiji M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale. 2019;11(18):8760–8775. doi:10.1039/c8nr09855g
26. Tonigold M, Simon J, Estupiñán D, et al. Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat Nanotechnol. 2018;13(9):862–869. doi:10.1038/s41565-018-0171-6
27. Sebak A, Gomaa I, ElMeshad A, et al. Distinct proteins in protein corona of nanoparticles represent a promising venue for endogenous targeting part I: in vitro release and uptake perspective. Int J Nanomed. 2020;15:8845–8862. doi:10.2147/IJN.S273713.
28. Sheikhsaran F, Sadeghpour H, Khalvati B, Entezar-Almahdi E, Dehshahri A. Tetraiodothyroacetic acid-conjugated polyethylenimine for integrin receptor-mediated delivery of the plasmid encoding IL-12 gene. Colloids Surf B Biointerfaces. 2017;150:426–436. doi:10.1016/j.colsurfb.2016.11.008
29. Sadeghpour H, Khalvati B, Entezar-Almahdi E, et al. Double domain polyethylenimine-based nanoparticles for integrin receptor mediated delivery of plasmid DNA. Sci Rep. 2018;8(1):1–12. doi:10.1038/s41598-018-25277-z
30. Mahmoudi R, Ashraf Mirahmadi-Babaheidri S, Delaviz H, et al. RGD peptide-mediated liposomal curcumin targeted delivery to breast cancer cells. J Biomater Appl. 2020;088532822094936. doi:10.1177/0885328220949367
31. Treuel L, Jiang X, Nienhaus GU. New views on cellular uptake and trafficking of manufactured nanoparticles. J R Soc Interface. 2013;10(82):20120939. doi:10.1098/rsif.2012.0939
32. Strobel C, Oehring H, Herrmann R, Förster M, Reller A, Hilger I. Fate of cerium dioxide nanoparticles in endothelial cells: exocytosis. J Nanoparticle Res. 2015;17:206. doi:10.1007/s11051-015-3007-4
33. Oh N, Park J-H-H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomedicine. 2014;9(SUPPL.1):51–63. doi:10.2147/IJN.S26592
34. Kettler K, Krystek P, Giannakou C, Hendriks AJ, Jong WHD. Exploring the effect of silver nanoparticle size and medium composition on uptake into pulmonary epithelial 16HBE14o-cells. J Nanoparticle Res. 2016;18(7):1–11. doi:10.1007/s11051-016-3493-z
35. Salvati A, Åberg C, Dos Santos T, et al. Experimental and theoretical comparison of intracellular import of polymeric nanoparticles and small molecules: toward models of uptake kinetics. Nanomed Nanotechnol Biol Med. 2011;7(6):818–826. doi:10.1016/j.nano.2011.03.005
36. Guarnieri D, Guaccio A, Fusco S, Netti PA. Effect of serum proteins on polystyrene nanoparticle uptake and intracellular trafficking in endothelial cells. J Nanoparticle Res. 2011;13:4295–4309. doi:10.1007/s11051-011-0375-2
37. Zhang Y, Huo M, Zhou J, Xie S. PKSolver: an add-in program for pharmacokinetic and pharmacodynamic data analysis in microsoft excel. Comput Methods Programs Biomed. 2010;99(3):306–314. doi:10.1016/j.cmpb.2010.01.007
38. Gunn J, Park SI, Veiseh O, Press OW, Zhang M. A pretargeted nanoparticle system for tumor cell labeling. Mol Biosyst. 2011;7(3):742–748. doi:10.1039/c005154c
39. Yin Y, Chen D, Qiao M, Wei X, Hu H. Lectin-conjugated PLGA nanoparticles loaded with thymopentin: ex vivo bioadhesion and in vivo biodistribution. J Control Release. 2007;123(1):27–38. doi:10.1016/j.jconrel.2007.06.024
40. Matsuo K, Itoh T, Koyama A, et al. CCR4 is critically involved in effective antitumor immunity in mice bearing intradermal B16 melanoma. Cancer Lett. 2016;378(1):16–22. doi:10.1016/j.canlet.2016.04.039
41. Bertrand N, Grenier P, Mahmoudi M, et al. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat Commun. 2017;8(1). doi:10.1038/s41467-017-00600-w
42. Rafiei P, Haddadi A. Docetaxel-loaded PLGA and PLGA-PEG nanoparticles for intravenous application: pharmacokinetics and biodistribution profile. Int J Nanomedicine. 2017;12:935–947. doi:10.2147/IJN.S121881
43. Kudgus RA, Walden CA, McGovern RM, Reid JM, Robertson JD, Mukherjee P. Tuning pharmacokinetics and biodistribution of a targeted drug delivery system through incorporation of a passive targeting component. Sci Rep. 2015;4(1):5669. doi:10.1038/srep05669
44. Mehrotra A, Kumar Pandit J. Preparation and characterization and biodistribution studies of lomustine loaded PLGA nanoparticles by interfacial deposition method. J Nanomedicine Biotherapeutic Discov. 2015;05(04). doi:10.4172/2155-983X.1000138
45. Shalgunov V, Zaytseva-zotova D, Zintchenko A, et al. Comprehensive study of the drug delivery properties of poly (L-lactide)-poly (ethylene glycol) nanoparticles in rats and tumor-bearing mice. J Control Release. 2017;261:31–42. doi:10.1016/j.jconrel.2017.06.006
46. Peñaloza JP, Márquez-Miranda V, Cabaña-Brunod M, et al. Intracellular trafficking and cellular uptake mechanisms of PHBV nanoparticles for targeted delivery in epithelial cell lines. J Nanobiotechnology. 2017;15(1). doi:10.1083/jcb.201002027
47. Cui Y, Song X, Li S, et al. The impact of receptor recycling on the exocytosis of Αvβ3 integrin targeted gold nanoparticles. Oncotarget. 2017;8(24):38618–38630. doi:10.18632/oncotarget.16955
48. Qiu L, Hu Q, Cheng L, et al. CRGDyK modified PH responsive nanoparticles for specific intracellular delivery of doxorubicin. Acta Biomater. 2016;30:285–298. doi:10.1016/j.actbio.2015.11.037
49. Sebak AA. Limitations of PEGylated nanocarriers: unfavourable physicochemical properties, biodistribution patterns and cellular and subcellular fates. Int J Appl Pharm. 2018;10(5):6–12. doi:10.22159/ijap.2018v10i5.27568
50. Alipour M, Baneshi M, Hosseinkhani S, et al. Recent progress in biomedical applications of RGD-based ligand: from precise cancer theranostics to biomaterial engineering: a systematic review. J Biomed Mater Res Part A. 2020;108(4):839–850. doi:10.1002/jbm.a.36862
51. Liu P, Sun Y, Wang Q, Sun Y, Li H, Duan Y. Intracellular trafficking and cellular uptake mechanism of MPEG-PLGA-PLL and MPEG-PLGA-PLL-gal nanoparticles for targeted delivery to hepatomas. Biomaterials. 2014;35(2):760–770. doi:10.1016/j.biomaterials.2013.10.020
52. Claudia M, Kristin Ö, Jennifer O, Eva R, Eleonore F. Comparison of fluorescence-based methods to determine nanoparticle uptake by phagocytes and non-phagocytic cells in vitro. Toxicology. 2017;378:25–36. doi:10.1016/j.tox.2017.01.001
53. Tedja R, Lim M, Amal R, Marquis C. Effects of serum adsorption on cellular uptake profile and consequent impact of titanium dioxide nanoparticles on human lung cell lines. ACS Nano. 2012;6(5):4083–4093. doi:10.1021/nn3004845
54. Reinholz J, Diesler C, Schöttler S, et al. Protein machineries defining pathways of nanocarrier exocytosis and transcytosis. Acta Biomater. 2018;71:432–443. doi:10.1016/j.actbio.2018.03.006
55. Hu L, Mao Z, Zhang Y, Gao C. Influences of size of silica particles on the cellular endocytosis, exocytosis and cell activity of HepG2 cells. J Nanosci Lett. 2011;1.
56. Sakhtianchi R, Minchin RF, Lee K-B, Alkilany AM, Serpooshan V, Mahmoudi M. Exocytosis of nanoparticles from cells: role in cellular retention and toxicity. Adv Colloid Interface Sci. 2013;201–202:18–29. doi:10.1016/j.cis.2013.10.013
57. Li Q, Xia D, Tao J, et al. Self-assembled core-shell-type lipid-polymer hybrid nanoparticles: intracellular trafficking and relevance for oral absorption. J Pharm Sci. 2017;106(10):3120–3130. doi:10.1016/j.xphs.2017.05.029
58. Panariti A, Miserocchi G, Rivolta I. The effect of nanoparticle uptake on cellular behavior: disrupting or enabling functions? Nanotechnol Sci Appl. 2012;5(1):87–100. doi:10.2147/NSA.S25515
59. Shang L, Nienhaus K, Nienhaus G. Engineered nanoparticles interacting with cells: size matters. J Nanobiotechnology. 2014;12(1):5. doi:10.1186/1477-3155-12-5
60. Salatin S, Yari Khosroushahi A. Overviews on the cellular uptake mechanism of polysaccharide colloidal nanoparticles. J Cell Mol Med. 2017;XX(X):1–19. doi:10.1111/jcmm.13110
61. Kettiger H, Schipanski A, Wick P, Huwyler J. Engineered nanomaterial uptake and tissue distribution: from cell to organism. Int J Nanomedicine. 2013;8(August):3255–3269. doi:10.2147/IJN.S49770
62. Guan X, Chen J, Hu Y, et al. Highly enhanced cancer immunotherapy by combining nanovaccine with hyaluronidase. Biomaterials. 2018;171:198–206. doi:10.1016/j.biomaterials.2018.04.039
63. Guo Y, Wang L, Lv P, Zhang P. Transferrin-conjugated doxorubicin-loaded lipid-coated nanoparticles for the targeting and therapy of lung cancer. Oncol Lett. 2015;9(3):1065–1072. doi:10.3892/ol.2014.2840
64. Wu M, Guo H, Liu L, Liu Y, Xie L. Size-dependent cellular uptake and localization profiles of silver nanoparticles. Int J Nanomedicine. 2019;14:4247–4259. doi:10.2147/IJN.S201107
65. Ekkapongpisit M, Giovia A, Follo C, Caputo G, Isidoro C. Biocompatibility, endocytosis, and intracellular trafficking of mesoporous silica and polystyrene nanoparticles in ovarian cancer cells: effects of size and surface charge groups. Int J Nanomedicine. 2012;7:4147–4158. doi:10.2147/IJN.S33803
66. Slowing II, Vivero-Escoto JL, Zhao Y, et al. Exocytosis of mesoporous silica nanoparticles from mammalian cells: from asymmetric cell-to-cell transfer to protein harvesting. Small. 2011;7(11):1526–1532. doi:10.1002/smll.201002077
67. Palchetti S, Pozzi D, Capriotti AL, et al. Influence of dynamic flow environment on nanoparticle-protein corona: from protein patterns to uptake in cancer cells. Colloids Surf B Biointerfaces. 2017;153:263–271. doi:10.1016/j.colsurfb.2017.02.037
68. Su G, Jiang H, Xu B, Yu Y, Chen X. Effects of protein corona on active and passive targeting of cyclic RGD peptide-functionalized PEGylation nanoparticles. Mol Pharm. 2018;15(11):5019–5030. doi:10.1021/acs.molpharmaceut.8b00612
69. Xiao W, Gao H. The impact of protein corona on the behavior and targeting capability of nanoparticle-based delivery system. Int J Pharm. 2018;552(1–2):328–339. doi:10.1016/j.ijpharm.2018.10.011
70. Ritz S, Schöttler S, Kotman N, et al. Protein corona of nanoparticles: distinct proteins regulate the cellular uptake. Biomacromolecules. 2015;16(4):1311–1321. doi:10.1021/acs.biomac.5b00108
71. Chen D, Ganesh S, Wang W, Amiji M. Plasma protein adsorption and biological identity of systemically administered nanoparticles. Nanomedicine (Lond). 2017;12(17):2113–2135. doi:10.2217/nnm-2017-0178
72. Palchetti S, Digiacomo L, Pozzi D, et al. Nanoparticles-cell association predicted by protein corona fingerprints †. Nanoscale. 2016;8:12755. doi:10.1039/c6nr03898k
73. Caracciolo G, Cardarelli F, Pozzi D, et al. Selective targeting capability acquired with a protein corona adsorbed on the surface of 1,2-dioleoyl-3-trimethylammonium propane/DNA nanoparticles. ACS Appl Mater Interfaces. 2013;5(24):13171–13179. doi:10.1021/am404171h
74. Rosso F, Marino G, Grimaldi A, et al. Vitronectin absorbed on nanoparticles mediate cell viability/proliferation and uptake by 3T3 Swiss Albino mouse fibroblasts: in vitro study. Biomed Res Int. 2013;2013:1–10. doi:10.1155/2013/539348
75. Gilkey MJ, Krishnan V, Scheetz L, Jia X, Rajasekaran A, Dhurjati P. Physiologically based pharmacokinetic modeling of fluorescently labeled block copolymer nanoparticles for controlled drug delivery in leukemia therapy. CPT Pharma Syst Pharmacol. 2015;4(3):e00013. doi:10.1002/psp4.13
76. Mohammad AK, Reineke JJ. Quantitative detection of PLGA nanoparticle degradation in tissues following intravenous administration. Mol Biol Cell. 2013;10:2183–2189. doi:10.1021/mp300559v
77. Godara S, Lather V, Kirthanashri SV, Awasthi R, Pandita D. Lipid-PLGA hybrid nanoparticles of paclitaxel: preparation, characterization, in vitro and in vivo evaluation. Mater Sci Eng C. 2020;109:110576. doi:10.1016/j.msec.2019.110576
78. Ahmed IS, El Hosary R, Hassan MA, Haider M, Abd-Rabo MM. Efficacy and safety profiles of oral atorvastatin-loaded nanoparticles: effect of size modulation on biodistribution. Mol Pharm. 2018;15(1):247–255. doi:10.1021/acs.molpharmaceut.7b00856
79. Alalaiwe A, Carpinone P, Alshahrani S, et al. Influence of chitosan coating on the oral bioavailability of gold nanoparticles in rats. Saudi Pharm J. 2019;27(2):171–175. doi:10.1016/j.jsps.2018.09.011
80. Wu C, Mu H. Lipid and PLGA microparticles for sustained delivery of protein and peptide drugs. PharmNanotechnol. 2019;07. doi:10.2174/2211738507666191029160944.
81. Navarro SM, Darensbourg C, Cross L, et al. Biodistribution of PLGA and PLGA/chitosan nanoparticles after repeat-dose oral delivery in F344 rats for 7 days. Ther Deliv. 2014;5(11):1191–1201. doi:10.4155/tde.14.79
82. Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–951. doi:10.1038/nbt.3330
83. Yu T, Hubbard D, Ray A, Ghandehari H. In vivo biodistribution and pharmacokinetics of silica nanoparticles as a function of geometry, porosity and surface characteristics. J Control Release. 2012;163:46–54. doi:10.1016/j.jconrel.2012.05.046
84. Jack Hu C-M, Kaushal S, Tran Cao HS, et al. Half-antibody functionalized lipid-polymer hybrid nanoparticles for targeted drug delivery to Carcinoembryonic Antigen (CEA) presenting pancreatic cancer cells. Mol Pharm. 2010;7(3):914–920. doi:10.1021/mp900316a
85. Palchetti S, Pozzi D, Mahmoudi M, Caracciolo G. Exploitation of nanoparticle-protein corona for emerging therapeutic and diagnostic applications. J Mater Chem B. 2016;4(25):4376–4381. doi:10.1039/c6tb01095d
86. Shen L, Tenzer S, Storck W, et al. Protein corona–mediated targeting of nanocarriers to B cells allows redirection of allergic immune responses. J Allergy Clin Immunol. 2018;142(5):1558–1570. doi:10.1016/j.jaci.2017.08.049
87. Nagayama S, Ogawara KI, Fukuoka Y, Higaki K, Kimura T. Time-dependent changes in opsonin amount associated on nanoparticles alter their hepatic uptake characteristics. Int J Pharm. 2007;342(1–2):215–221. doi:10.1016/j.ijpharm.2007.04.036
88. Mirshafiee V, Kim R, Park S, Mahmoudi M, Kraft ML. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials. 2016;75:295–304. doi:10.1016/j.biomaterials.2015.10.019
89. Schöttler S, Klein K, Landfester K, Mailänder V. Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake. Nanoscale. 2016;8(10):5526–5536. doi:10.1039/c5nr08196c
90. Bros M, Nuhn L, Simon J, et al. The protein corona as a confounding variable of nanoparticle-mediated targeted vaccine delivery. Front Immunol. 2018;9. doi:10.3389/fimmu.2018.01760.
91. Schöttler S, Becker G, Winzen S, et al. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat Nanotechnol. 2016;11(4):372–377. doi:10.1038/nnano.2015.330
92. Aoyama M, Hata K, Higashisaka K, Nagano K, Yoshioka Y, Tsutsumi Y. Clusterin in the protein corona plays a key role in the stealth effect of nanoparticles against phagocytes. Biochem Biophys Res Commun. 2016;480(4):690–695. doi:10.1016/j.bbrc.2016.10.121
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.