Back to Journals » OncoTargets and Therapy » Volume 10
Panel of seven long noncoding RNA as a candidate prognostic biomarker for ovarian cancer
Authors Zhan XH, Dong C, Liu G, Li YX , Liu L
Received 26 November 2016
Accepted for publication 28 December 2016
Published 1 June 2017 Volume 2017:10 Pages 2805—2813
DOI https://doi.org/10.2147/OTT.S128797
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tohru Yamada
Xiaohui Zhan,1,2 Chuanpeng Dong,3 Gang Liu,3 Yixue Li,1,2,4 Lei Liu3
1Key Lab of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, 2University of Chinese Academy of Sciences, Beijing, 3Institute of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai, 4Shanghai Center for Bioinformation Technology, Shanghai Industrial Technology Institute, Shanghai, People’s Republic of China
Abstract: Ovarian cancer is one of the most common and lethal gynecological malignancies. The diagnosis of ovarian cancer is often at an advanced stage. Accumulated evidence suggests that long noncoding RNAs (lncRNAs) play important roles during ovarian tumorigenesis. In this study, using the lncRNA-mining approach, we analyzed lncRNA expression profiles of 493 ovarian cancer patients from Gene Expression Omnibus datasets, and identified a signature group of seven lncRNAs (BC037530, AK021924, AK094536, AK094536, BC062365, BC004123 and BC007937) associated with patient survival in the training dataset GSE9891. We also formulated a risk score model to divide patients into low-risk and high-risk groups based on the expression of these seven lncRNAs. We further validated the predictive power of our risk score model in two other datasets, GSE26193 and GSE63885. Our analysis showed that the seven-lncRNA signature can serve as an independent predictor apart from Federation of Gynecology and Obstetrics (FIGO) stage and patient age. Further investigation revealed the seven-lncRNA signature correlated with few critical signaling pathways involved in cancer. Combined, all these findings strongly support that the seven-lncRNA signature can serve as a strong prognosis biomarker.
Keywords: lncRNA, ovarian cancer, prognostic, gene signature, survival
Introduction
Ovarian cancer is one of the most common and lethal gynecological malignancies.1 The 5-year survival rate of patients with early-stage (Federation of Gynecology and Obstetrics [FIGO] stage I) is >90%, whereas the 5-year survival rate of patients with advanced-stage (FIGO stages III and IV) plunges to <30%.2 The poor prognosis is mainly due to the usually indistinct symptoms, lack of reliable screening test and tumor resistance to chemotherapy.3 Therefore, there is strong need to identify effective prognostic biomarkers to help optimize and personalize treatment.4,5
Long noncoding RNAs (lncRNAs) are defined as RNAs that are longer than 200 nucleotides and have little or no protein-coding ability.6 Most lncRNAs are expressed in specific tissues and specific cancer types.7 Despite lncRNAs having no protein-coding ability, a great number of studies have demonstrated that lncRNAs participate in diverse biological processes, such as development,8 differentiation,9 energy metabolism,10 apoptosis,11 angiogenesis.12 LncRNAs can influence almost every step of the life cycle for gene regulation, and play important roles in cancers development.13,14
Despite the importance of lncRNAs in carcinogenesis and development, relevant studies on lncRNA as possible prognostic biomarkers for ovarian cancer are still limited. Large-scale lncRNAs data produced recently make such studies possible. In this study, we aimed to identify lncRNAs expression signatures that can predict ovarian cancer patient survival. Using the publicly released ovarian cancer dataset (GSE9891)15 from the Gene Expression Omnibus (GEO) database, we identified a signature including seven lncRNAs associating with survival, and proposed a risk score formula based on their expressions to predict the patient survival. The prognostic risk score model was further validated in the cohorts GSE26193 and GSE63885.16,17 Our findings indicated that the seven-lncRNA signature can serve as a strong prognostic biomarker for ovarian cancer.
Materials and methods
Microarray processing and lncRNA profile mining
The three lncRNA gene expression data (CEL files) and corresponding clinical data of ovarian cancer in this study were downloaded from the GEO database and processed using Robust Multichip Average (RMA) algorithm for background adjustment. We used GATExplorer software to annotate the lncRNA microarray probes.18 LncRNA mapper was obtained from GATExplorer to calculate RNA expression. We only included the lncRNA probes that are mapped to human genome and mouse genome (derived from RNA database [RNAdb]).19 For the microarray expression analysis, we threshold the lncRNAs included at least minimum of three probes mapping in the corresponding noncoding RNAs (ncRNAs) entity.
Gene Set Enrichment Analysis (GSEA)
GSEA was performed upon 186 curated Kyoto Encyclopedia of Genes and Genomes gene sets by the GSEA, which was developed by Broad Institute (Cambridge, MA, USA) and the gene sets provided by MSigDB (Molecular Signatures Database) were considered as reference.20 Gene sets with a false discovery rate (FDR) value <0.05 after performing 1,000 permutations were considered to be significantly enriched.
Statistical analysis
Univariable Cox proportional hazards regression analysis was carried out in the training set (GSE9891) to assess the association between lncRNA gene expression and patient survival information. lncRNAs with P<0.001, FDR <0.001 were selected to be strongly correlated with patient survival. Random survival forests variable hunting (RSFVH) algorithm was carried out to select predictors.21 Using the previously selected genes fitted in a multivariable Cox regression model, a risk score formula was constructed based on the expression of these lncRNAs to predict the patient survival. Each patient had a risk score and the risk score was the weighted combination of Cox regression coefficients for each significant lncRNAs expression.22,23 The patients were divided into two groups (ie, low-risk group and high-risk group) using the median risk score as the cutoff. The Kaplan–Meier method was used to estimate the survival time and the two-sided log rank test was used to compare the survival difference between the low-risk and high-risk groups.
Furthermore, we used Cox multivariate analysis to test whether the risk score model was independent of patient age and FIGO Stage with available data. The receiver operating characteristic (ROC) curves were also used to compare the sensitivity and specificity of the lncRNA risk score in predicting survival in respect to FIGO Stage and patient age.24 All above analyses were accomplished by R program.
Results
Preanalysis of GEO ovarian cancer gene expression data
The lncRNA gene expression data and corresponding clinical data of ovarian cancer in this study were downloaded from the GEO database. The selected data sets included >100 patients with corresponding survival information. We analyzed the correlation between lncRNA expression signatures and survival endpoints for ovarian cancer as a whole (overall survival [OS], progress-free survival [PFS] and disease-free survival [DFS]). Our strategy was to use the largest data set (GSE9891, n=285)15 as a training set to identify the lncRNA expression signature. Then another two smaller independent datasets (GSE26193, n=107; GSE63885, n=101)16,17 were used as testing sets in this study. After removing samples without corresponding clinical survival information, a total of 460 samples were left, 278 of which were from GSE9891, 107 from GSE26193 and 75 from GSE63885. Figure 1 describes the workflow of this study.
Identification of prognostic lncRNA genes from the training set
In order to identify the prognostic lncRNA genes, we used univariable Cox proportional hazards regression to analysis lncRNA expression data in training set. Here we identified a set of 33 lncRNAs strongly correlated with patients’ overall survival (P<0.001) from a total of 5,635 lncRNAs. Considering smaller panel of lncRNA will make a more practical model, the RSFVH algorithm was used to select the more relevant genes, and consequently a set of seven-lncRNA genes (BC037530, AK021924, AK094536, BC062365, AK130460, BC007937 and BC004123) were finally identified (Figure 2).21,25 Among these lncRNAs, BC037530 has the highest relevant importance value in the seven predictors. Expression of all seven-lncRNAs gene was strongly correlated with patient survival (Table 1). The lncRNAs (BC037530, AK021924, AK094536, BC062365 and AK130460) with the positive coefficients indicated that higher expressions of these lncRNAs were associated with poor survival, while the lncRNAs (BC007937 and BC004123) with the negative coefficients indicated that lower expressions of these lncRNA were associated with poor survival.
  | Figure 2 Random survival forests variable hunting analysis. |
An seven-lncRNA signature predicts the survival of ovarian cancer patients in the training set
In order to investigate how well the seven-lncRNA signature could predict the survival of ovarian cancer patients, we constructed a risk score formula based on the expression of these seven lncRNAs to predict the patient survival in the training set. Each patient had a risk score, and the risk score was a weighted combination of Cox regression coefficients for all seven significant lncRNAs. The risk scores based on the seven-lncRNA expression were calculated as follows:
|
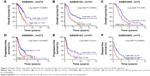
Using the median risk score as cutoff, we divided the patients into two groups (low-risk group, n=139 and high-risk group, n=139). Patients in the high-risk group had a shorter survival time than patients in the low-risk group (log-rank test P<0.0001, median survival time 24 vs 32 months; Figure 3A). The correlation of the risk score and survival was also significant when it was evaluated as a continuous variable in the univariable Cox regression model. The distribution of patient risk scores, survival status and lncRNA values were analyzed independently for the training set (Figure 4). We found that patients with high-risk scores tended to have higher expressions of BC037530, AK021924, AK094536, AK094536, BC062365 and lower expressions of BC004123, BC007937.
Validation of the seven-lncRNA signature for survival prediction in the testing sets
In order to test whether the seven-lncRNA signature as ovarian cancer survival predictors overfit the data, we used another two independent lncRNA datasets: GSE26193 (n=107) and GSE63885 (n=75), for validation. As with the pre-analysis method of the training set, we carried out the same pre-analysis for the testing set. The OS, PFS and DFS were evaluated respectively. By using the same cutoff selecting method as the training set, we got a consistent result with the training set, supporting that the lncRNA signature is a robust predictor for survival. Patients in the high-risk group had a significantly shorter survival time than patients in the low-risk group (Figure 3B–F) indicating the robustness of our model.
Multivariate regression analysis shows that the seven-lncRNA expression signature is independent of age/grade and stage
Studies have proved that age at diagnosis, FIGO stage, grade of tumor were significant (P≤0.05) prognostic factors for overall survival of ovarian cancer.26 Cox multivariate analysis was performed to check whether the seven-lncRNA expression signature was an independent predictor of ovarian cancer patient’s survival. We used multivariate Cox proportional hazard model to evaluated whether age, grade and stage as covariates had effect on the of ovarian cancer patient survival. The results showed that for OS analysis, our lncRNA signature was independent of age/grade and stage in GSE9891 and GSE26193. For PFS/DFS analysis, the signature was independent of age/grand and stage in GSE9891, GSE26193 and GSE63885. In summary, the risk score based on the expression of the seven lncRNAs may serve as an independent predictor for ovarian cancer patient survival (Tables 2 and 3).
Evaluation of the risk score performance by ROC curve analysis
We performed ROC analysis in the training set to assess the sensitivity and specificity of survival prediction between our model and FIGO stage and patient age. By comparing the areas under ROC (AUROC) of ROC curves, we found that our risk score model was better than FIGO stage and patient age to predict the patient survival. The AUROC of seven-lncRNA gene signature, FIGO stage signature and patient age signature were 0.742, 0.626 and 0.534, respectively. We also observed significant difference between our model with FIGO stage (P=0.0006) and our model with patient age (P<0.0001; Figure 5). This indicated that our model was more sensitive and specific than the existing model in predicting the survival of ovarian cancer patients.
Identification of biological pathways and processes associated with the seven lncRNAs
GSEA was performed to identify associated biological processes and signaling pathways. We compared the gene expression profiles of patients with high-risk and low-risk groups and found that extracellular matrix (ECM)–receptor interaction, transforming growth factor (TGF)-beta signaling pathway and focal adhesion activity were enriched in the high-risk group (Figure 6).
Discussion
In this study, we have identified a seven-lncRNA signature panel (BC037530, AK021924, AK094536, BC062365, AK130460, BC007937 and BC004123), which was associated with the ovarian cancer patient survival from the training set. We also constructed a prognostic risk score model based on the seven-lncRNA expression signature, patients with high-risk scores tended to have a shorter survival time than patients with low-risk scores. The model was future validated in two other independent lncRNA datasets (GSE26193 and GSE63885), and further investigation revealed that the seven-lncRNA signature may well used to predict the ovarian cancer patient survival time.
We used the multivariable Cox regression analysis with age/grand and stage as covariables to evaluate whether the seven-lncRNA signature was an independent biomarker. The results showed that the seven-lncRNA expression signature was independent biomarker adjusted by age/grade and stage in GSE9891 (OS/PFS), GSE63885 (OS/PFS), except for GSE26193, which only exhibited significance with the univariate Cox analysis. This may be due to the smaller sample dataset or some other unknown confounding factors employed, the P-value was not significant for the OS analysis in GSE26193. Taken together, these results still suggested that the seven-lncRNA signature functions as an independent predictor of ovarian cancer patient survival. Besides that, ROC analysis also indicating that the seven-lncRNA risk score model was comparable with the currently main prognostic models: FIGO stage and patient age. All these indicting that the seven-lncRNA signature might be a well survival predictor no matter in the technical point or in the clinical point.
Unfortunately, functional studies of these genes lncRNAs in cancer have not been reported so far. Nevertheless, our study demonstrated associations of these lncRNAs with patient survival. GSEA proved that the seven-lncRNA signature was more likely associated with ECM–receptor interaction, TGF-beta signaling pathway, focal adhesion. ECM–receptor interaction plays an important role in invasion and metastasis of tumor.27 TGF-beta signaling pathway is critical for proliferation, differentiation, development and apoptosis of tumor.28 Focal adhesion also functions in invasion and metastasis of tumor cells.29 It is conceivable that the overexpression or downregulation of these lncRNAs in those pathways can affect the course of carcinogenesis.
Limitation of this study should be acknowledged as well. First, we only included 5,635 (out of 15,000+) human lncRNAs in the present study. The lncRNA signature as a prognostic biomarker may not represent all the lncRNA candidates that were potentially correlated with the overall survival of ovarian cancer patient. Second, the functions of these seven lncRNAs in tumorigenesis were still unknown, even though the associated biological processes and signaling pathways were inferred. Finally, even through the seven-lncRNAs signature was tested as prognostic biomarker in two other datasets, experimental studies such as PCR, clinical trials and the function analysis were still needed. Because there are still possibilities of false positives in selection of the seven lncRNAs for the prediction of clinical outcome.
In summary, we have identified a prognostic seven-lncRNA panel and demonstrated it as a powerful prognostic predictor for ovarian cancer. Based on the expression of these seven lncRNAs, a risk score model was established to predict the poor and good prognosis cases. Further multivariate Cox regression analysis revealed that the seven-lncRNA signature was independent of the main prognostic factors-American Joint Committee on Cancer (AJCC) stage, age/grade and might help personalize prediction of ovarian cancer prognosis. Moreover, these lncRNAs may involved in biological processes such as ECM–receptor interaction, TGF-beta signaling pathway, focal adhesion, which are all known key players in tumorgenesis. Despite some drawbacks, our seven-lncRNA signature can potentially serve as a powerful prognostic marker for ovarian cancer.
Acknowledgments
This work was supported by the National Key Research and Development Program on Precision Medicine (2016YFC0901700, 2016YFC0901900 and 2016YFC0901600), the National Key Technology Support Program (2013BAI101B09) and the National High Technology Research and Development Program of China (2015AA020104 and 2015AA020108).
Author contributions
All authors conceived and designed the project. Xiaohui Zhan, Chuanpeng Dong, and Gang Liu collected and analyzed the data. Xiaohui Zhan drafted the manuscript. All authors contributed toward critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Cho KR, Shih I. Ovarian cancer. Annu Rev Pathol. 2009;4:287–315. | ||
Fishman DA, Bozorgi K. The scientific basis of early detection of epithelial ovarian cancer: the national ovarian cancer early detection program (Nocedp). In: Stack MS, Fishman DA, editors. Ovarian Cancer. New York, NY: Springer US; 2002:3–28. | ||
Langhe R. microRNA and ovarian cancer. Adv Exp Med Biol. 2015;889:119–151. | ||
Au KK, Josahkian JA, Francis JA, Squire JA, Koti M. Current state of biomarkers in ovarian cancer prognosis. Future Oncol. 2015;11(23):3187–3195. | ||
Wei W, Dizon D, Vathipadiekal V, Birrer MJ. Symposium article Ovarian cancer: genomic analysis. Ann Oncol. 2013;24 (Suppl 10):x7–x15. | ||
Mendell JT. Clinical implications of basic research targeting a long noncoding RNA in breast cancer. N Engl J Med. 2016;374:2287–2289. | ||
Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. | ||
Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. | ||
Spurlock CF 3rd, Tossberg JT, Guo Y, Collier SP, Crooke PS 3rd, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. 2015;6:6932. | ||
Rupaimoole R, Lee J, Haemmerle M, et al. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13(11):2395–2402. | ||
Su S, Liu J, He K, et al. Overexpression of the long noncoding RNA TUG1 protects against cold-induced injury of mouse livers by inhibiting apoptosis and inflammation. FEBS J. 2016;283(7):1261–1274. | ||
Fu WM, Lu YF, Hu BG, et al. Long noncoding RNA hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7(4):4712–4723. | ||
Gan L, Xu M, Zhang Y, Zhang X, Guo W. Focusing on long noncoding RNA dysregulation in gastric cancer. Tumour Biol. 2015;36(1):129–141. | ||
Schmitt AM, Chang HY. Perspective long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. | ||
Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14(16):5198–5208. | ||
Mateescu B, Batista L, Cardon M, et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17(12):1627–1636. | ||
Lisowska KM, Olbryt M, Dudaladava V, et al. Gene expression analysis in ovarian cancer – faults and hints from DNA microarray study. Front Oncol. 2014;4:6. | ||
Risueño A, Fontanillo C, Dinger ME, De Las Rivas J. GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs. BMC Bioinformatics. 2010;11:221. | ||
Pang KC, Stephen S, Engström PG, et al. RNAdb–a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:125–130. | ||
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. | ||
Ishwaran H, Kogalur UB. Consistency of Random Survival Forests. Stat Probab Lett. 2010;80(13–14):1056–1064. | ||
Alizadeh AA, Gentles AJ, Alencar AJ, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118(5):1350–1358. | ||
Bralten LB, French PJ. Genetic alterations in glioma. Cancers (Basel). 2011;3(1):1129–1140. | ||
Kang J, D’Andrea AD, Kozono D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J Natl Cancer Inst. 2012;104(9):670–681. | ||
Kawaguchi A, Iwadate Y, Komohara Y, et al. Gene expression signature-based prognostic risk score in patients with primary central nervous system lymphoma. Clin Cancer Res. 2012;18(20):5672–5681. | ||
Wu XS, Wang XA, Wu WG, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15(6):806–814. | ||
Rejniak KA, editor. Systems Biology of Tumor Microenvironment. Quantitative Modeling and Simulations (Vol. 936). Cham, Switzerland: Springer Nature; 2016. | ||
Kamato D, Burch ML, Piva TJ, et al. Transforming growth factor-β signalling: role and consequences of Smad linker region phosphorylation. Cell Signal. 2017;25(10):2017–2024. | ||
Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.