Back to Journals » International Journal of General Medicine » Volume 16
Pan-Cancer Analysis of the Tumorigenic Effect and Prognostic Diagnostic Value of FAM111B in Human Carcinomas
Received 16 March 2023
Accepted for publication 11 May 2023
Published 16 May 2023 Volume 2023:16 Pages 1845—1865
DOI https://doi.org/10.2147/IJGM.S409690
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Hengmiao Wu, Chao Liang
Department of General Surgery, the Affiliated Lihuili Hospital, Ningbo University, Ningbo, 315000, People’s Republic of China
Correspondence: Chao Liang, Department of General Surgery, the Affiliated Lihuili Hospital, Ningbo University, 57 Xingning Road, Ningbo, 315000, People’s Republic of China, Tel +86-574-87018607, Email [email protected]
Introduction: FAM111B (FAM111 trypsin-like peptidase B) gene mutations have been linked to a hereditary fibrosing poikiloderma disorder known to cause poikiloderma, tendon contracture, myopathy, and pulmonary fibrosis (POIKTMP). Overexpression of FAM111B has been associated with an increased risk of certain cancers with a poor prognosis, although the relationship between FAM111B and other tumors is still unclear, and the molecular mechanism of its action is not fully understood.
Methods: We investigated the biological functions of FAM111B in 33 solid tumors using multi-omics data. We further recruited 109 gastric cancer (GC) patients for a clinical cohort study to confirm the effect of FAM111B on early tumor recurrence. Furthermore, we assessed the role of FAM111B in GC cell proliferation and migration via EdU incorporation, CCK8 and transwell assays in vitro.
Results: We found that FAM111B can enhance oncogenesis and progression in multiple tumor types. The clinical cohort of GC showed that upregulation of FAM111B is associated with early recurrence of GC, and knockdown of the FAM111B gene can inhibit the proliferation and migration of GC cells. Gene enrichment analysis indicates that FAM111B promotes cancer through immune system process, chromosome instability, DNA repair, and apoptosis regulation. Mechanistically, FAM111B appears to promote the growth cycle of malignant tumor cells while inhibiting apoptosis.
Conclusion: FAM111B may serve as a potential pan-cancer biomarker for predicting the prognosis and survival of malignant tumor patients. Our study elucidates the role of FAM111B in the occurrence and development of various cancers, and highlights the need for future research on FAM111B in cancers.
Keywords: FAM111B, pan-cancer, biomarker, prognosis, survival
Introduction
Malignant tumors are one of the leading causes of death worldwide, and the global morbidity and burden of cancer are increasing.1 Despite significant progress in modern cancer therapies, such as immunotherapy and radiotherapy, a considerable number of patients continue to advance to later stages of cancer, resulting in poor outcomes.2 The lack of definite tumor diagnostic markers is an important reason for this poor prognosis.3 Therefore, it is essential to find appropriate tumor markers through pan-cancer analysis and analyze their relationship with prognosis.
FAM111 trypsin-like peptidase B (FAM111B), encodes a C-terminal protein that contains a pancreatitis cysteine/serine peptide domain. Mutations in FAM111B are associated with a rare genetic disorder known as hereditary fibrous disease, poikiloderma with tendon contractures, myopathy, and pulmonary fibrosis (POIKTMP).3–5 Moreover, two cases of pancreatic cancer have been linked to FAM111B mutations,6 suggesting that these mutations not only contribute to the development of POIKTMP but may also lead to cancer formation.7 The exact mechanism by which FAM111B mutations contribute to tumorigenesis remains unclear. However, some reports suggest that FAM111B participates in the p53 signaling pathway, promoting the progression of lung adenocarcinoma (LUAD).8 Another study reported that FAM111B enhances the proliferation and cell cycle progression of LUAD cells by degrading p16, a tumor suppressor protein.9 These findings proposes that FAM111B could be a novel target for treating this type of lung cancer. Furthermore, high expression of FAM111B has been linked to poor prognosis in breast cancer, where it is believed to play a critical role in cell proliferation, migration, and invasion.10 The evidence suggests that FAM111B may function as an oncogene, given its association with poor survival across multiple cancer types. However, most research on FAM111B has been limited to specific cancer types, and there has been no comprehensive pan-cancer analysis of the link between FAM111B and other cancers.
In this study, we aim to conduct a pan-cancer analysis of FAM111B, utilizing various datasets such as TCGA, GEO, and GTEx. Through integration of gene expression, survival prognosis, genetic alteration, immune infiltration, and gene enrichment data of FAM111B across multiple malignancies, in conjunction with clinical cohorts of gastric cancer (GC) patients and in vitro experiment of GC cell lines, our results revealed the significance of etiology and clinical prognostic implications of FAM111B in cancer.
Materials and Methods
Data Collection and Expression Analysis
We obtained RNA-seq data for the FAM111B gene from the University of California, Santa Cruz (UCSC) Genome Browser.11 To investigate differences in FAM111B expression between tumors and adjacent normal tissues, we utilized Tumor Immunity Estimation Resource (TIMER) 2.0 with data from TCGA.12 In cases where limited or no normal tissue was available in TIMER 2.0, we further assessed expression differences using Gene Expression Profiling Interaction Analysis (GEPIA).13 Additionally, we analyzed FAM111B expression levels at each pathological stage in all TCGA cancers using GEPIA2. Moreover, we conducted an analysis of the Clinical Proteomic Tumor Analysis Consortium (CPTAC) dataset using the UALCAN website to examine and contrast FAM111B protein expression levels in normal and primary tumor tissues across the six tumor types.14
To assess the expression level of FAM111B in GC, tumor tissues and corresponding paracancerous tissues were obtained from 109 patients with primary GC who underwent surgery at Lihuili Hospital Affiliated to Ningbo University (China) between December 2020 and December 2022. Clinical information for these samples was also collected. All experimental protocols were approved by the Ethics Committee of Lihuili Hospital Affiliated to Ningbo University (KY2022SL436-01). The ethics committee approved the waiver of informed consent due to the retrospective nature of the review. All participant data had been anonymized or kept confidential and complied with the Declaration of Helsinki. The two-year disease-free survival was determined by measuring the time elapsed between the date of resection and the date of tumor recurrence.
Cell Line
Human GC cell lines (AGS, MKN-45) were purchased from the Shanghai Haixing biological company. AGS and MKN-45 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin at 37°C under 5% CO2.
Cell Transfection
2×105 GC cells were plated into 6-well plates and transfected with a scrambled negative control siRNA (NC-siRNA), FAM111B-siRNA (FAM111B-Homo-1052, 5′-GUGCCACUGAUGAAAUUAATT-3′, si-1; FAM111B-Homo-1336, 5′- GAGGCAAUUAAUCUCUUAATT-3′, si-2; FAM111B-Homo-1541, 5′-GCGAACAGCUUACAUAUUATT −3′, si-3) (all purchased from GenePharma) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 6~8 h, the medium (Opti-MEM, Gibco) was replaced with complete culture medium. The transfected GC cells were incubated in different groups for the indicated time periods. All experiments were performed in 48 h after transfection.
RNA Extraction and qPCR Analysis
According to the previous method,15 total RNA was extracted from GC tissues and GC cell lines using Trizol reagent (Invitrogen). Subsequently, qRT-PCR was performed using the PrimeScript RT Master Mix (TaKaRa) and TB Green Premix Ex Taq (TaKaRa) by the LightCycler 480 (Roche). The relative mRNA expression levels were calculated by the 2−ΔΔCt method. The primers used were as follows: FAM111B: forward: GCCCTTGAAATGCAGAATCCA and reverse: GCTGTAAACACACTACGGTCTAA; and GAPDH: forward: GTCGGAGTCAACGGA TTTGG and reverse: CGGTGCCA TGGAA TTTGCC.
Western Blot Analysis
After 72 hours of cell transfection, total protein was extracted and the concentration was measured using the previously described method.15 The lysates were loaded onto SDS-PAGE for electrophoresis and then transferred onto a PVDF membrane. The membrane was blocked with 5% skim milk for 1 hour and incubated with primary antibodies against FAM111B (Proteintech, dilution 1:2000) and GAPDH (CST, dilution 1:4000) at 4 °C overnight. After washing with Tris-buffered saline with Tween 20 three times, a secondary goat anti-rabbit antibody (CST, dilution 1:4000) was added to the membrane and incubated for 1 hour. Protein bands were visualized using enhanced chemiluminescence.
Cell Viability Assay
Cell viability was measured using Cell Counting Kit-8 (CCK8, Dojindo) according to the manufacturer’s instructions. 5000 GC cells were plated into 96-well plates for the indicated time periods. After incubation with CCK-8 solution for 3 h, the absorbance of the wells was measured at 450 nm using an MRX II microplate reader (Dynex). Relative cell viability was calculated as a percentage of negative control GC cells. Cell proliferation assays were performed using the Click-iT 5-ethynyl-20-deoxyuridine (EdU) Imaging Kit (Invitrogen) according to the manufacturer’s instructions. After transfection, GC cells were seeded into 96-well plates at a density of 10,000 cells per well. Cells were incubated with medium containing EdU for 2 h. Subsequently, cells were fixed with 4% paraformaldehyde for 30 minutes and permeabilized with 0.5% Triton-X 100 for 20 min. The reaction mixture was added and incubated for 30 min in the dark. After washing with PBS, cells were stained with DAPI for 5 min. The number of EdU-positive cells was measured using fluorescence microscopy and ImageJ software.
Cell Migration Assay
The transfected cells were resuspended in serum-free medium at a concentration of 5 × 104/mL. A total of 100 µL of cell suspension was added to the upper compartment of transwell assay inserts (8-mm pores; Corning) containing 200 μL of serum-free DMEM. The lower compartment was filled with 800 µL DMEM containing 10% FBS. After incubating at 37°C for 24 h, the chamber was removed, and the medium was aspirated. Cells on the upper surface of the filter were removed by wiping with a cotton swab, and those that had migrated through the pores and adhered to the lower surface of the filter were fixed with methanol for 30 min, stained with crystal violet for 15 min and imaged using microscopy. The migration ability was evaluated by counting the number of stained cells in five randomly selected fields per filter in each group.
Prognosis—survival Analysis
We used GEPIA2 to determine the overall survival (OS) and disease-free survival (DFS) of FAM111B in all TCGA tumors.13 High cutoff (50%) and low cutoff (50%) were used to distinguish between the high and low expression groups. Hypothesis testing was performed using log-rank analysis, and survival plots were generated via GEPIA2. Next, we used the “pROC” software program to acquire TCGA and GTEx data and generate receiver operating characteristic (ROC) curves. The ROC curve’s abscissa and ordinate denote the false positive rate and true positive rate, respectively. A larger area under curve (AUC) under the ROC curve indicates a more accurate prediction.
Genetic Alteration Analysis
We utilized the cBioPortal website to obtain information on the characterization of the FAM111B genetic alteration.16 Through this platform, we analyzed the frequency and type of FAM111B alterations, as well as copy number alterations (CAN), in all TCGA tumors. Additionally, we were able to view the mutation site information of FAM111B in all TCGA tumors.
Immune Infiltration Analysis
We investigated the association between FAM111B expression and immune infiltration in all TCGA tumors using the TIMER2 web server. We centered on two immune cell types - CD8+ T cells and cancer-associated fibroblasts (CAFs) - and assessed immune infiltration employing diverse algorithms. In order to adjust for tumor purity, we employed the purity-adjusted Spearman rank correlation test to compute p-values and partial correlation values.
Related Gene Enrichment Analysis
We utilized several tools and databases to explore FAM111B co-expression, gene regulation, and protein interactions. Firstly, the STRING program was used to generate a co-expression network of Homo sapiens FAM111B.17 Next, we extracted 100 FAM111B-related genes from the most similar transcription factors to FAM111B from the TCGA dataset using GEPIA2. These genes were subjected to Gene Ontology (GO) pathway enrichment analysis using Metascape.18 We generated the FAM111B-protein interaction network using BioGRID.19 Furthermore, we performed pairwise gene regression analysis via GEPIA2.
Statistical Analysis
We conducted all statistical analyses using R (version 4.0.3). The statistical analyses involved the use of different tests depending on the distribution of the variables. Specifically, the t-test, one-way ANOVA and two-way ANOVA were employed to analyze normally distributed variables, while nonparametric tests were used for non-normally distributed variables. Correlation analysis was conducted using Pearson’s correlation and Spearman’s rank correlation test. A significance level of P < 0.05 was used to determine statistical significance.
Results
Differential Expression of FAM111B Between Human Normal and Cancerous Tissues
This study was designed to evaluate the tumorigenic role of FAM111B in humans. We analyzed the expression levels of FAM111B in all tumors and adjacent normal tissues of TCGA via TIMER2. The level of FAM111B expression in the cancer tissues of bladder urothelial carcinoma (BLCA), head and neck squamous cell carcinoma (HNSC), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), cholangiocarcinoma (CHOL), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), LUAD, lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), stomach adenocarcinoma (STAD) (P < 0.001), rectum adenocarcinoma (READ), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (P < 0.01), pheochromocytoma and paraganglioma (PCPG) (P < 0.05) is lower than the adjacent normal tissues (Figure 1A).
We collected normal tissue data from the GTEx database as a control to evaluate the difference in FAM111B expression between tumor and paracancer and normal tissues of uterine carcinosarcoma (UCS), diffuse large B cell lymphoma (DLBC), thymoma (THYM), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), acute myeloid leukemia (LAML), glioblastoma multiforme (GBM), and esophageal carcinoma (ESCA) (P < 0.05) (Figure 1B). We observed that the expression of FAM111B in both paracancerous and normal tissues was lower than that in tumor tissues, although the difference was not significant in some tumors, including adrenocortical carcinoma (ACC), testicular germ cell tumors (TGCT) and brain lower grade glioma (LGG).
Based on data obtained from the CPTAC database, it was found that the expression of the FAM111B protein was significantly elevated in primary tumor tissues of UCEC, HNSC (P < 0.001), LUAD (P < 0.01), and ovarian cancer (P < 0.05) when compared to normal tissues. However, this pattern was not observed in breast cancer and GEM (Figure 1C).
Furthermore, the relationship between FAM111B expression and various pathological stages of different cancers was investigated using GEPIA2. We observed significant differences in ACC, KICH, KIRP, LIHC, SKCM, LUAD, THCA (all P < 0.05) (Figure 2A) but not in others (Figure 2B). These findings suggest that FAM111B expression may be associated with the progression of certain types of cancer.
Prognostic Value of FAM111B in Diverse Human Cancers
We conducted an analysis of the relationship between FAM111B expression and the prognosis of patients with various cancers from the TCGA database, who were divided into two groups according to high or low expression of FAM111B. The study revealed a positive correlation between high expression of FAM111B and poor OS in patients with ACC, KIRP, LGG, LIHC, LUAD, MESO and PEAD (all P < 0.05). Conversely, low expression of FAM111B was associated with poor OS in READ, THCA and THYM (all P < 0.05) (Figure 3A).
Similarly, we found a significant correlation between high expression of FAM111B and poor DFS in patients with several cancer types, including ACC, BLCA, KIRP, LGG, LIHC, PEAD and THCA (all P < 0.05). Conversely, low expression of FAM111B was associated with poor DFS in STAD (P < 0.05) (Figure 3B).
As shown in Figure 4, we found that FAM111B has high accuracy in predicting the majority of tumors, indicating that FAM111B can be utilized as a general biomarker for tumor prediction.
 |
Figure 4 Analysis of ROC curves between FAM111B and tumor prediction based on data from TCGA and GTEx databases. |
Genetic Alteration Analysis of FAM111B in Pan-Cancer
The genetic variation status of FAM111B in various cancers within the TCGA database (consisting of 10,967 samples across 32 studies) was obtained using the cBioPortal online site. Results showed that FAM111B mutations were most frequently (>4%) observed in UCEC tumor samples, with the majority of these mutations being missense mutations. Additionally, “deep deletion” was identified as the predominant type in MESO, with an altered frequency of approximately 2% (Figure 5A and B). In total, 127 FAM111B mutations were detected across all TCGA samples, including 108 missense mutations, 17 truncating mutations, and 2 sv/fusion mutations (Figure 5C). The mutation site A513V was found to be the most common.
Correlation of FAM111B Expression Levels with Tumor Immune Microenvironment
In advanced stages of some tumor types, tumor-related immune infiltration may become nonviable or even interact with tumor cells to promote cancer progression and metastasis.20,21 Unfortunately, many patients exhibit innate or acquired resistance to immunotherapy. CAFs are known to secrete a diverse array of cytokines and hinder the penetration of drugs or therapeutic immune cells into tumor tissues, which diminishes the effectiveness of tumor immunotherapy.22,23 Multiple algorithms were used to evaluate the correlation between the expression of FAM111B and the level of immune infiltration in various TCGA cancers. The results obtained from the multiple algorithmic analyses demonstrated a statistically significant negative correlation between FAM111B expression and the estimates of CAFs infiltration in BRCA, STAD, and THYM cancers (P < 0.05, Figure 6A). Conversely, a positive correlation was observed between the infiltration of CD8+ T cells and the expression of FAM111B in KIRC, HNSC-HPV, THYM, and UVM cancers, as indicated by most algorithms (P < 0.05, Figure 7A). To illustrate our findings, we generated scatterplots of these cancers using the most efficient algorithm (Figures 6B and 7B). In conclusion, the expression of FAM111B is associated with immune infiltration in some cancers.
Correlations of FAM111B Expression Levels with Tumor Heterogeneity
Tumor heterogeneity significantly influences the initiation, progression, and response to therapy of tumors. Tumor mutational burden (TMB) and microsatellite instability (MSI) are two biomarkers that are closely associated with tumor heterogeneity. Elevated levels of TMB and MSI are typically linked with greater tumor heterogeneity and improved immunotherapy outcomes.24,25 Our results demonstrated a significant correlation between FAM111B expression and TMB in 15 cancers, with 14 of them being positively correlated, such as GBMLGG, LGG and LUAD, while THYM showed a negative correlation (Figure 8A and B). In addition, we detected a positive correlation between FAM111B and MSI for MESO and ACC, but negative for DLBC, GBMLGG. Mutant-allele tumor heterogeneity (MATH) effectively represented tumor specificity, and we observed a significant correlation with it in 12 tumors, with a significant positive correlation in 6 tumors, such as LUAD, LAML and BRCA, and a significant negative correlation in 6 tumors, such as GBMLGG, LGG, and KIRC (Figure 8C). Tumor purity and ploidy are two crucial parameters that reflect tumor heterogeneity, and play a significant role in modulating the efficacy of tumor therapy. We found a positive correlation between FAM111B expression and purity in most cancers (Figure 8D), but an inconsistent correlation in tumors where we found a negative correlation between ploidy and FAM111B expression (Figure 8E). Homologous recombination deficiency (HRD) status, which is a key prognostic indicator for multiple tumor treatment strategies and outcomes, showed a significant correlation with FAM111B expression in 17 cancers, including GEM, LGG, and LUAD, as shown in Figure 8F. Neoantigen (NEO) are novel antigens generated as a result of mutations in tumor cells that can be recognized as foreign by the immune system, and represent a potential target for cancer immunotherapy. Our findings reveal a significant correlation between NEO and FAM111B in TGCT, suggesting that FAM111B may serve as a potential factor promoting the generation of NEO in TGCT (Figure 8G). We also observed a significant correlation between FAM111B expression and loss of heterozygosity (LOH) in various cancers, with most showing a positive correlation (Figure 8H).
Enrichment Analysis of FAM111B-Related Genes
To explore the mechanism of FAM111B in cancer development, we used GEPIA2 to identify the top 100 genes in the TCGA dataset with expression patterns comparable to FAM111B (Supplementary Table S1). Next, we used the Metascape website to enrich the functions of these genes and found that they were closely related to immune system process, chromosome instability and DNA repair (Figure 9A). We further validated these results by using the STRING tool to obtain 50 proteins that were co-expressed with FAM111B (Figure 9B). These findings suggest that FAM111B may be involved in these biological processes by interacting with key proteins involved in chromosome instability, DNA repair, and regulation of apoptosis. The interaction network was exhibited in Figure 8C, which is also illustrated it by the analysis based on the GeneMANIA website (Figure 9C). According to the BioGRID 4.3 database, FAM111B physically interacts with SET, HIST1H1E, MED, ITGB1, EXOSC9, DTX2, BAG3 and ATP13A2 (Figure 9D). These proteins function in immune system process, chromosome instability, DNA repair and apoptosis regulation.26–29 The correlation analysis demonstrated robust positive associations between the expression of FAM111B and SET, HIST1H1E, EXOSC9, DTX2, BAG3, and ATP13A2, as depicted in a scatter plot (Figures 9E–J). Based on these findings, we hypothesize that FAM111B may promote tumor development by impacting processes related to immune system process, chromosome instability, DNA repair and apoptosis regulation.
Clinical Cohort Validation of Gastric Cancer
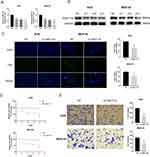
In the previous study, we found that the expression of FAM111B in GC tissue was increased compared with that in paracancerous tissue, but the patients with high expression of FAM111B in TCGA-STAD had significantly higher DFS time than those with low expression. To validate the clinical relevance of FAM111B in GC prognosis, we examined the expression of FAM111B in 109 pairs of tumor tissue and paracancerous normal tissue samples from GC patients. The qRT-PCR analysis showed that FAM111B was upregulated in tumor tissue compared to paracancerous normal tissue (Figure 10A). We further analyzed the relationship between FAM111B and recurrence in GC patients. GC patients (n = 109) were separated into two groups by the median FAM111B expression level. As shown in Figure 10B, patients with lower FAM111B expression (n = 55) showed longer recurrence-free survival (RFS) than patients with higher FAM111B expression (n = 54). Further analysis of the correlation between FAM111B expression and clinicopathological parameters showed significant differences in stage T and tumor grade between the high and low FAM111B expression groups (Table 1). In univariate analyses, expression levels of FAM111B and node metastasis and stage T were identified as significant prognostic factors for RFS (Table 2). However, in multivariate analyses, only FAM111B expression level was identified as a prognostic factor for RFS (Table 3). These results indicated that FAM111B represents a biomarker of early GC recurrence.
 |
Table 1 Correlation Between FAM111B Expression and GC Clinicopathological Parameters |
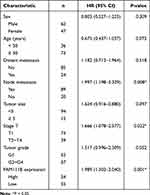 |
Table 2 Univariate Analyses of Prognostic Factors for Recurrence-Free Survival |
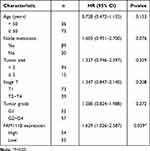 |
Table 3 Multivariate Analyses of Prognostic Factors for Recurrence-Free Survival |
Effects of FAM111B Knockdown on Gastric Cancer Cell Lines
To further confirm the role of the FAM111B gene in GC cell lines, we employed small interfering RNA (siRNA) to knockdown the expression level of FAM111B in GC cell lines, and qRT-PCR and Western blot experiments were used to test the knockdown efficiency (Figure 11A and B). The group si-3 that exhibited the highest efficiency of FAM111B knockdown was selected for subsequent experiments. EDU and CCK8 assays demonstrated that the suppression of FAM111B significantly impeded the proliferation of GC cells (Figure 11C and D). Furthermore, we verified the effect of FAM111B silencing on the migration of gastric cancer cells through transwell experiments (Figure 11E). Our findings suggest that knockdown of FAM111B can affect the proliferation and migration of GC cells.
Discussion
Previous studies have revealed mutations FAM111B gene in the human are frequently associated with a rare multisystem fibrotic disease, POIKTMP,4,30 progressive osseous heteroplasia, and mutations of unknown genetic polymorphism.30,31 Accumulating evidence in recent years has shown that FAM111B mutation is associated with increased susceptibility to malignancy.7,32 Overexpression of FAM111B in cancer cells has been associated with accelerated cancer progression and poor clinical outcomes.8,9 It is still unknown if FAM111B can regulate the development of various malignancies through known or unidentified molecular mechanisms. FAM111B has not yet been the subject of a pan-cancer analysis that has been reported in published literature. Therefore, using a variety of datasets, including as TCGA, GEO, CPTAC, and others, we thoroughly assessed the FAM111B1 gene in all cancers. Analysis was done on the molecular traits, survival outlook, diagnostic utility, gene mutation, immunological infiltration, signaling pathways, and gene enrichment.
We discovered that the FAM111B gene is significantly expressed in most TCGA cancer types and there are already articles verifying its promoting effect in different tumors, such as pancreatic cancer and breast cancer.6,10 Another study utilizing in vitro functional analysis of FAM111B knockout cells demonstrated that FAM111B plays an important role in the proliferation and cell cycle progression of KRAS-driven LUAD under serum starvation, and that FAM111B expression is significantly associated with clinical progression.9 The outcomes of our genetic differential analysis are congruent with prior findings indicating that FAM111B is overexpressed in LUAD, breast cancer, and PAAD. Furthermore, the correlation between FAM111B expression and clinical variables or prognosis is also in line with previous reports. High FAM111B expression is linked to a poor prognosis in various tumor types, such as LUAD and PAAD. Moreover, we verified the significance of FAM111B in GC through 109 clinical cohorts. The expression of FAM111B is significantly abundant in GC tissue than in paracancerous tissue, which is consistent with the TCGA-STAD samples. However, the RFS of the clinical cohort exhibited disparities compared to that of the TCGA-STAD samples, which requires further exploration. Cell proliferation and migration abilities are important indicators for evaluating the prognosis of GC patients. Furthermore, we conducted cell experiments and observed that the knockdown of FAM111B inhibited the proliferation and migration of GC cells in vitro.
Next, we investigated the potential association between FAM111B expression and immune infiltration in various cancers within TCGA. Utilizing multiple algorithms, we found statistically significant correlations between FAM111B expression and cancer-associated fibroblast infiltration in BRCA, STAD, THYM, and TGCT. Similarly, we observed statistically significant correlations between FAM111B expression and CD8+ T cell immune infiltration in HNSC-HPV+, KIRC, THYM, and UVM cancers. Tumor heterogeneity, which arises from differences in tumor cell morphology, genetics, and biochemistry, is closely tied to a tumor’s degree of malignancy and can have a significant impact on tumor diagnosis and treatment.30,33 To further investigate the relationship between FAM111B and tumor heterogeneity, we performed an analysis of eight different indicators including TMB, MATH, and MSI. TMB and MSI are two crucial molecular biological characteristics that can be used to predict the response to immune checkpoint inhibitor therapy in certain cancer patients. Thus, our analysis of pan-cancer heterogeneity of FAM111B may provide a novel reference for predicting the response and prognosis of immunotherapy across various cancers.
To further explore the specific mechanisms of FAM111B in cancer, gene enrichment analysis revealed that FAM111B is closely related to DNA repair and immune system processes, which is consistent with previous study.32 Using STRING and GeneMANIA database, we found numerous genes that co-express and physically interact with FAM111B in tumors, such as SET, HIST1H1E, EXOSC9, DTX2, BAG3, and ATP13A2. These genes primarily encode proteins that are involved in DNA repair, apoptosis regulation, and cell cycle regulation.26–29 These results are consistent with the outcomes of gene enrichment studies related to FAM111B, indicating a potential molecular mechanism by which FAM111B could be involved in cancer progression. We have confirmed the reduction of DNA replication activity in gastric cancer cells after knockdown of FAM111B through the EdU assay. Recent studies have demonstrated that FAM111B mutations are associated with cell cycle dysregulation in POIKTMP.34 And in the field of tumors, it was also found that FAM111B is also implicated in the regulation of cell cycle and its related signaling pathways.35 Notably, in some cancer types, FAM111B knockout tumor cells have shown a reduction in the expression of anti-apoptotic genes, including BCL-2 and BAG3, which is consistent with our findings regarding the association between FAM111B and apoptosis genes.8,36 Based on these findings, we propose that the abnormal expression of FAM111B and its non-specific degradation of DNA-related proteins lead to genome instability, disrupt DNA repair processes, and cause cell cycle dysregulation, ultimately contributing to the development, progression, and susceptibility of cancer.
Nonetheless, the current study has certain limitations that warrant consideration. Firstly, bioinformatics analysis depends on public databases or datasets provided by other researchers. However, variations in experimental conditions, analysis methods, or software package selection may result in different results, which may affect the reproducibility of bioinformatics analysis. Secondly, bioinformatics analysis can uncover the expression patterns of genes or proteins. Although we have validated these findings in a clinical cohort of GC and in vitro experiments, subsequent in vivo experiments are required to confirm their biological significance. Thirdly, the FAM111B gene is not a heavily studied gene, and limited attention has been given to certain types of cancers, resulting in a paucity of data and restricted sample sizes, which may negatively impact the results.
Conclusion
In conclusion, our study results indicate that FAM111B may serve as a potential prognostic biomarker for several types of cancers, validated through clinical validation in a GC cohort and in vitro experiments. Moreover, our bioinformatics analyses revealed that FAM111B is associated with immune infiltration, DNA repair, and apoptotic regulation, providing potential mechanisms underlying analysis of immune infiltration and gene enrichment of FAM111B. These results provide initial insights into the function and relevance of FAM111B in cancer development and pave the way for future prospective investigations.
Data Sharing Statement
This research recruited public databases and website tools. The original data and R codes can be obtained from the author for reasonable requests.
Ethics Approval
All experimental protocols were approved by the Ethics Committee of Lihuili Hospital Affiliated to Ningbo University (KY2022SL436-01). The ethics committee approved the waiver of informed consent due to the retrospective nature of the review. All participant data had been anonymized or kept confidential and complied with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Funds of Young Scientists of China [grant number:81802944]; Program of Ningbo Medical Research Center for Digestive System Cancer [grant number:2019A21003]; Key Project of Ningbo Natural Science Foundation [grant number: 2021J279].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Chen W, Li Y, Guo L, Zhang C, Tang S. Long non-coding RNA FTX predicts a poor prognosis of human cancers: a meta-analysis. Biosci Rep. 2021;41(1). doi:10.1042/BSR20203995
3. Roshani M, Baniebrahimi G, Mousavi M, et al. Exosomal long non-coding RNAs: novel molecules in gastrointestinal cancers’ progression and diagnosis. Front Oncol. 2022;12:1014949. doi:10.3389/fonc.2022.1014949
4. Mercier S, Kury S, Shaboodien G, et al. Mutations in FAM111B cause hereditary fibrosing poikiloderma with tendon contracture, myopathy, and pulmonary fibrosis. Am J Hum Genet. 2013;93(6):1100–1107. doi:10.1016/j.ajhg.2013.10.013
5. Wu Y, Wen L, Wang P, Wang X, Zhang G. FAM111BCase Report: diverse phenotypes of congenital poikiloderma associated with mutations in codon 628: a case report and literature review. Front Genet. 2022;13:926451. doi:10.3389/fgene.2022.926451
6. Mercier S, Kury S, Nahon S, Salort-Campana E, Barbarot S, Bezieau S. FAM111B mutation is associated with pancreatic cancer predisposition. Pancreas. 2019;48(5):e41–e42. doi:10.1097/MPA.0000000000001303
7. Welter A, Machida Y. Functions and evolution of FAM111 serine proteases. Front Mol Biosci. 2022;9:1081166. doi:10.3389/fmolb.2022.1081166
8. Sun H, Liu K, Huang J, et al. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. Onco Targets Ther. 2019;12:2829–2842. doi:10.2147/OTT.S190934
9. Kawasaki K, Nojima S, Hijiki S, et al. FAM111B enhances proliferation of KRAS-driven lung adenocarcinoma by degrading p16. Cancer Sci. 2020;111(7):2635–2646. doi:10.1111/cas.14483
10. Li W, Hu S, Han Z, Jiang X. YY1-induced transcriptional activation of FAM111B contributes to the malignancy of breast cancer. Clin Breast Cancer. 2022;22(4):e417–e25. doi:10.1016/j.clbc.2021.10.008
11. Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi:10.1038/s41587-020-0546-8
12. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W14. doi:10.1093/nar/gkaa407
13. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W60. doi:10.1093/nar/gkz430
14. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
15. Dong ZB, Wu HM, He YC, et al. MiRNA-124-3p.1 sensitizes hepatocellular carcinoma cells to sorafenib by regulating FOXO3a by targeting AKT2 and SIRT1. Cell Death Dis. 2022;13(1):35. doi:10.1038/s41419-021-04491-0
16. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
17. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D68. doi:10.1093/nar/gkw937
18. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
19. Oughtred R, Rust J, Chang C, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30(1):187–200. doi:10.1002/pro.3978
20. Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi:10.1038/nature10673
21. Kikuchi T, Mimura K, Ashizawa M, et al. Characterization of tumor-infiltrating immune cells in relation to microbiota in colorectal cancers. Cancer Immunol Immunother. 2020;69(1):23–32. doi:10.1007/s00262-019-02433-6
22. Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nature reviews. Cancer. 2020;20(3):174–186. doi:10.1038/s41568-019-0238-1
23. Desbois M, Wang Y. Cancer-associated fibroblasts: key players in shaping the tumor immune microenvironment. Immunol Rev. 2021;302(1):241–258. doi:10.1111/imr.12982
24. Chan T, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. doi:10.1093/annonc/mdy495
25. Luchini C, Bibeau F, Ligtenberg M, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. 2019;30(8):1232–1243. doi:10.1093/annonc/mdz116
26. Flex E, Martinelli S, Van Dijck A, et al. Aberrant function of the C-terminal tail of HIST1H1E accelerates cellular senescence and causes premature aging. Am J Hum Genet. 2019;105(3):493–508. doi:10.1016/j.ajhg.2019.07.007
27. Yang L, Zhang Q, Niu T, Liu H. SET levels contribute to cohesion fatigue. Mol Biol Cell. 2021;32(13):1256–1266. doi:10.1091/mbc.E20-12-0778
28. Ciolfi A, Aref-Eshghi E, Pizzi S, et al. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin Epigenetics. 2020;12(1):1–11. doi:10.1186/s13148-019-0804-0
29. Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco M. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2(4):e141–e41. doi:10.1038/cddis.2011.24
30. Arowolo A, Rhoda C, Khumalo N. Mutations within the putative protease domain of the human FAM111B gene may predict disease severity and poor prognosis: a review of POIKTMP cases. Exp Dermatol. 2022;31(5):648–654. doi:10.1111/exd.14537
31. Pan X, Zheng S. Clinical and genetic characteristics of nevus of Ota with choroidal melanoma in Chinese. Ophthalmic Genet. 2019;40(4):338–341. doi:10.1080/13816810.2019.1650073
32. Goussot R, Prasad M, Stoetzel C, Lenormand C, Dollfus H, Lipsker D. Expanding phenotype of hereditary fibrosing poikiloderma with tendon contractures, myopathy, and pulmonary fibrosis caused by FAM111B mutations: report of an additional family raising the question of cancer predisposition and a short review of early-onset poikiloderma. JAAD Case Rep. 2017;3(2):143–150. doi:10.1016/j.jdcr.2017.01.002
33. Pe’er D, Ogawa S, Elhanani O, Keren L, Oliver TG, Wedge D. Tumor heterogeneity. Cancer Cell. 2021;39(8):1015–1017. doi:10.1016/j.ccell.2021.07.009
34. Rhoda C, Sunda F, Kidzeru E, Khumalo NP, Arowolo A. FAM111B dysregulation promotes malignancy in fibrosarcoma and POIKTMP and a low-cost method for its mutation screening. Cancer Treat Res Commun. 2023;34:100679. doi:10.1016/j.ctarc.2022.100679
35. Dereure O. Poïkilodermie fibrosante héréditaire avec contractures, myopathie et fibrose pulmonaire: responsabilités de mutations de FAM111B [Hereditary fibrosing poikiloderma with contractures, myopathy and pulmonary fibrosis: The role of mutations in FAM111B]. Ann Dermatol Venereol. 2014;141(6–7):478–479. French. doi:10.1016/j.annder.2014.03.017
36. Hoffmann S, Pentakota S, Mund A, et al. FAM111 protease activity undermines cellular fitness and is amplified by gain-of-function mutations in human disease. EMBO Rep. 2020;21(10):e50662. doi:10.15252/embr.202050662
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.