Back to Journals » Cancer Management and Research » Volume 12
Paeonol Inhibits Cell Proliferation, Migration and Invasion and Induces Apoptosis in Hepatocellular Carcinoma by Regulating miR-21-5p/KLF6 Axis
Authors Cai M, Shao W, Yu H, Hong Y, Shi L
Received 18 March 2020
Accepted for publication 24 June 2020
Published 17 July 2020 Volume 2020:12 Pages 5931—5943
DOI https://doi.org/10.2147/CMAR.S254485
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Sanjeev K. Srivastava
Miaoguo Cai,1,* Wei Shao,1,* Huijun Yu,2 Ye Hong,1 Lili Shi3
1Department of Medical Oncology, Luqiao Branch of Taizhou Hospital, Taizhou City, Zhejiang Province, People’s Republic of China; 2Department of Pediatric, Luqiao Branch of Taizhou Hospital, Taizhou City, Zhejiang Province, People’s Republic of China; 3Department of Infection, Luqiao Branch of Taizhou Hospital, Taizhou City, Zhejiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Shao
Department of Medical Oncology, Luqiao Branch of Taizhou Hospital, No. 1 Xialiqiao West Road, Luqiao District, Taizhou City 318050, Zhejiang Province, People’s Republic of China
Tel +86-57682421047
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is one of the most common tumors with high mortality. MicroRNAs (miRNAs) were reported as crucial markers for the diagnosis of HCC. Paeonol exerted many pharmacological effects on tumor progression. This study aimed to elucidate the underlying molecular mechanism of paeonol in HCC progression.
Methods: Cell viability was determined by Cell Counting Kit-8 (CCK-8) assay. Cell apoptosis was examined by flow cytometry. The levels of Cyclin D1, cyclin-dependent kinase 4 (CDK4), B-cell lymphoma-2 (Bcl-2) and Bcl-2 associated X protein (Bax) were detected by Western blot assay. Cell migration and invasion were assessed by transwell assay. The levels of matrix metalloproteinase-2 (MMP2) and matrix metalloproteinase-9 (MMP9) were measured by Western blot. The expression of miR-21-5p and kruppel-like factor 6 (KLF6) was detected by quantitative real-time PCR (qRT-PCR) or Western blot assay, respectively. Dual-luciferase reporter assay was performed to analyze the interaction between miR-21-5p and KLF6. The enrichment of miR-21-5p was determined by RNA pull-down assay. Xenograft assay was conducted to analyze tumor growth in vivo.
Results: The results demonstrated that cell viability of Hep3B and Huh-7 cells was inhibited, while cell apoptosis was promoted after treatment with paeonol. Transwell assay indicated that cell migration and invasion were blocked in paeonol-treated cells. Moreover, miR-21-5p expression was markedly decreased in paeonol-treated cells and its knockdown suppressed cell viability, migration and invasion, but contributed to cell apoptosis. MiR-21-5p targeted KLF6 and its silencing prominently elevated KLF6 level. Furthermore, the restoration experiment determined that miR-21-5p and KLF6 were antagonisms on cell viability, apoptosis, migration and invasion. Also, paeonol abated the decrease in KLF6 level caused by miR-21-5p up-regulation. Besides, paeonol suppressed tumor growth in vivo.
Conclusion: Paeonol impeded cell viability, migration and invasion and triggered apoptosis by regulating miR-21-5p/KLF6 axis in HCC cells. Xenograft assay confirmed that paeonol inhibited tumor growth through miR-21-5p/KLF6 axis in HCC in vivo.
Keywords: hepatocellular carcinoma, paeonol, miR-21-5p, KLF6
Introduction
Liver is a metabolic organ of the body and plays the role of detoxification, storage of liver sugar, and synthesis of secretory proteins.1 Liver diseases, including hepatitis, cirrhosis, hepatapostema and hepatocellular carcinoma, are regarded as major obstacles to the health of people worldwide.2 Hepatocellular carcinoma (HCC) is one of the most common cancers in the world,3,4 causing about 750,000 deaths worldwide each year,5 and China accounts for half of all deaths. Limited treatment choices are the main cause of high mortality.6 As previously reported, there are many factors that contributed to HCC, such as alcohol drinking, hepatic toxin, hepatitis virus infection, and gene modification.7 According to the Barcelona-Clinic-Liver-Cancer (BCLC) staging system, HCC patients usually go through four stages: early subclinical stage, subclinical stage, metaphase stage and late stage.8 The stage of cancer directly determines the mortality rate of HCC patients. Early HCC patients can even be completely cured by hepatectomy or liver transplantation.9 For patients with HCC for more than five years, the survival rate was less than 14%.10 Hence, the diagnostic biomarkers for early HCC are urgently needed for the treatment and prevention of HCC.
Paeonol (2ʹ-hydroxy-4ʹ-methoxyacetophenone) is extracted from the herb Pycnostelma paniculatum K.S and Paeonia suffruticosa Andrews.11 In recent years, paeonol has attracted worldwide attention due to its antineoplastic activity. In vitro experiments demonstrated that paeonol could cause the death of malignant tumor cells.12 Also, paeonol plays a vital role as an anti-inflammatory, anti-oxidant agent in HCC.13,14 Although the pharmacological activity of paeonol has been extensively proposed, the mechanism of paeonol in the progression of HCC remains unclear.
MicroRNAs (miRNAs) are a family of non-coding RNAs and act as vital regulators in many physiological and biological processes, including cell viability, apoptosis, migration and invasion.15–17 Mature miRNAs regulate gene expression by the complementary match with the 3ʹ-untranslated region (3ʹ-UTR) of the target mRNA.18 A large number of miRNAs were identified to be involved in the diagnosis and therapy of HCC.19–21 MiR-21-5p, as a crucial regulatory factor in tumorigenesis, was proved to play a potential role in metabolism and apoptosis of HCC.22 The previous study indicated that miR-21 up-regulation enhanced disease progression in HCC patients.23 Moreover, the aberrant expression of miR-21-5p made it a prognostic biomarker for HCC.24,25 Moreover, miR-21 promoted cell migration and invasion via regulating KLF5 in HCC.26 However, the molecular mechanism of miR-21-5p in HCC needs to be further investigated.
Kruppel-like factor 6 (KLF6) is a member of the sp1/KLF transcription factor family and acts as a tumor suppressor gene in human cancers. KLF6 is widely expressed in human tissues and plays a pivotal role in cell cycle, apoptosis and differentiation.27 He et al reported that KLF6 blocked cell proliferation and cell cycle in HCC cells.28 Besides, KLF6 expression was significantly down-regulated in HCC.29 These data implied that KLF6 was associated with HCC progression and might be an essential target for HCC treatment. Nevertheless, the function of KLF6 in paeonol-treated HCC cells and the network of paeonol/miR-21-5p/KLF6 in HCC remain further research.
Our research was designed to explore the role of paeonol in HCC and the molecular mechanism of paeonol in cell viability, apoptosis, migration and invasion in vitro and in xenograft model in vivo.
Materials and Methods
Cell Culture and Paeonol Treatment
HCC cell lines (Hep3B and Huh-7) were provided by the Institute of Basic Medical Sciences of Peking Union Medical College (Beijing, China). Cells were maintained in Dulbecco’s Modified Eagle Medium (Gibco, Carlsbad, CA, USA) supplementing with 10% fetal bovine serum (Hyclone, Logan, UT, USA). All cells were cultured in a constant temperature incubator at 37°C with 5% CO2. Paeonol was purchased from the National Institutes for Food and Drug Control (Beijing, China). For different concentrations of paeonol (50 µM, 100 µM, 200 µM, 400 µM, 800 µM), varying amounts of normal saline were used to dissolve paeonol. HCC cell lines were cultivated in medium with various concentrations of paeonol for 24 h.
Cell Transfection
MiR-21-5p inhibitor, miR-21-5p mimic and their corresponding negative controls (NC inhibitor and NC mimic), small interference RNA (si-RNA) against KLF6 (si-KLF6) and si-RNA against NC (si-NC) were all obtained from Genepharma Co. Ltd (Genepharma, Shanghai, China). These oligos were transfected into Hep3B and Huh-7 cells with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). After incubation for 24 h, cells were collected for further experiment.
Cell Viability Assay
Six groups of Hep3B cells and six groups of Huh-7 cells were prepared in 96-well plates, each well containing approximately 10,000 cells. Six concentrations of paeonol (0 µM, 50 µM, 100 µM, 200 µM, 400 µM, 800 µM) were transferred into cells, respectively. Then, the cells treated with paeonol were cultured for 24 h at 37°C with 5% CO2. Subsequently, 10 µL of cell counting kit-8 (CCK-8) solution (Beyotime, Shanghai, China) was added into cells. After incubation for 4 h, the absorbance was detected by a microplate reader (Bio-Rad, Hercules, CA, USA) at 450 nm. The value of absorbance is proportional to the number of cells.
Flow cytometry. For cell apoptosis assay, the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) was used to analyze cell apoptosis. HCC cells were harvested after treatment with paeonol or miRNA inhibitor for 1 day. The cells were then resuspended with binding buffer and 5 μL Annexin V-FITC and PI solution. Dark incubation for 10 min, the apoptosis cells were determined by flow cytometer (BD Biosciences). For cell cycle assay, Hep3B and Huh-7 cells were incubated with paeonol (50 µM or 800 µM). Then, cells were resuspended in 200 μL PBS containing 10 μL PI and incubated at room temperature in the dark for 15 min. The cell cycle was monitored by the flow cytometer (BD Biosciences) with CellQuest Software (Becton–Dickinson).
Western Blot Assay
The cells were lysed in lysis buffer (Beyotime) and then subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel electrophoresis to separate proteins by molecular weight. Next, the proteins were blotted onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Then, the membranes were treated with 5% non-fat milk for 1 h. Next, the membranes were incubated with primary antibodies against Cyclin D1 (ab16663; 1/100; Abcam, Cambridge, MA, USA), CDK4 (ab108357; 1/5000; Abcam), Bcl-2 (ab32124; 1/1000; Abcam), Bax (ab32503; 1/4000; Abcam), MMP2 (ab37150; 1 µg/mL; Abcam), MMP9 (ab38898; 1/1000; Abcam), KLF6 (ab241385; 1/1000; Abcam) and GAPDH (ab8245; 1/2000; Abcam) overnight at 4°C. Next, the membranes were interacted with horseradish peroxidase-conjugated secondary antibody (ab6721; Abcam) with 1/3000 dilution at room temperature for 2 h. Finally, the protein bands were visualized by enhanced chemiluminescence chromogenic substrate (Beyotime) and measured by Image Lab software (Bio-Rad).
Cell Migration and Invasion Assays
Cell migration and invasion were detected by transwell assays. First, about 40,000 cells were seeded in the upper chambers (Corning, New York, NY, USA) and incubated at 37°C for 24 h. Then, the cells through the membrane were incubated with 0.5% crystal violet (Sigma, St. Louis, MO, USA). Finally, a microscope (Olympus, Tokyo, Japan) was used to observe the migrated and invaded cells. For cell invasion assay, the upper chamber was coated by Matrigel (Becton Dickinson, Franklin Lakes, NJ, USA) prior to inoculation, and cell migration assay was not required.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA of Hep3B and Huh-7 cells was extracted by RNA extraction Kit (TransGen Biotech, Beijing, China). TransScript miRNA First-Strand cDNA Synthesis Kit (TransGen Biotech) was utilized for the synthesis of qRT-PCR templates. TransStart Top Green qPCR SuperMix (TransGen) was used to perform qRT-PCR. The primers for miR-21-5p and internal control were purchased from Sangon Biotech (Sangon, Shanghai, China). All data were calculated by 2−ΔΔCt method. The sequences of primers for miR-21-5p and U6 small RNA were listed as follows: U6: Forward: 5ʹ-GCGCGTCGTGAAGCGTTC-3ʹ, Reverse: 5ʹ-GTGCAGGGTCCGAGGT-3ʹ; miR-21-5p: Forward: 5ʹ-TGCGCTAGCTTATCAGACTGAT-3ʹ, Reverse: 5ʹ-CCAGTGCAGGGTCCGAGGTATT-3ʹ.
Dual-Luciferase Reporter Assay
TargetScan online database was used to predict the downstream gene of miR-21-5p. To confirm the relationship between miR-21-5p and KLF6, the sequences of KLF6 3ʹ-UTR harboring wt-KLF6 or mut-KLF6 were amplified and cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA), respectively. The luciferase reporter plasmids were constructed by Hanbio (Hanbio, Shanghai, China). Hep3B and Huh-7 cells were co-transfected with miR-21-5p mimic or NC mimic and wt-KLF6 or mut-KLF6 plasmids, and the luciferase activity was determined by dual-luciferase reporter kit (Promega). All experiments were repeated at least three times.
RNA Pull-Down Assay
Biotin-labeled negative control (Bio-NC) and KLF6 (Bio-KLF6) were purchased from Sangon (Sangon). Hep3B and Huh-7 cells were cultivated at 37°C overnight and then transfected with Bio-KLF6 or Bio-NC for 8 h. After transfection, cells were washed by PBS buffer and lysed using cell lysis buffer. The Streptavidin-Dyna beads were added into cell lysates to isolate RNA bound to Bio-KLF6. Finally, miR-21-5p enrichment in Hep3B and Huh-7 cells was detected by qRT-PCR.
Xenograft Experiment
BALB/c mice (5 weeks) were randomly divided into three groups (n=6 per group). Hep3B cells (2×104) were subcutaneously injected into the right-back of the mice. Mice in the drug treatment group were intragastrically injected with paeonol (5 mg/kg or 80 mg/kg) every day.30 Tumor volume was measured every 7 days. After 4 weeks, the mice were sacrificed by cervical dislocation. If spontaneous breathing and blink reflex were not observed within 2–3 min, the mice were considered dead. Then, the excised xenografts were weighed. The xenograft assay was authorized by the Ethical Committee of the Institution for Animal Experimentation and the Animal Welfare Committee of Luqiao Branch of Taizhou Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
All data were validated by three independent biological replicates and calculated with the means ± standard deviation (SD). Student’s t-test for two groups and one-way analysis of variance (ANOVA) for at least three groups were performed to analyze the differences. All data were arranged by the GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). The P-value less than 0.05 was considered as statistically significant.
Results
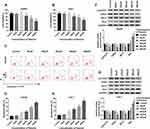
Paeonol Inhibited Cell Viability and Induced Apoptosis in Hep3B and Huh-7 Cells
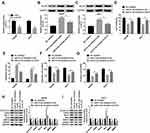
To investigate the potential role of paeonol in HCC, CCK-8 assay was performed to detect cell viability. After treatment with varying concentrations of paeonol (0 μM, 50 μM, 100 μM, 200 μM, 400 μM, 800 μM), cell viability of Hep3B and Huh-7 cells was substantially decreased in a dose-dependent manner (Figure 1A and B). Moreover, the cell apoptosis rate in Hep3B and Huh-7 cells was gradually increased with the growing concentration of paeonol (Figure 1C–E). In addition, the expression levels of proteins associated with cell proliferation or apoptosis were evaluated by Western blot assay. With the increasing concentration of paeonol, Cyclin D1, CDK4 and Bcl-2 levels were apparently blocked, whereas Bax level was obviously promoted in Hep3B and Huh-7 cells (Figure 1F and G). Taken together, paeonol could suppress cell viability and enhance apoptosis in Hep3B and Huh-7 cells.
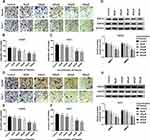
Paeonol Blocked Cell Migration and Invasion of Hep3B and Huh-7 Cells
To further confirm the function of paeonol in HCC, transwell assay was carried out to examine cell migration and invasion. Paeonol treatment remarkably inhibited HCC cell proliferation at 36 h (Supplement Figure 1A and B), so we performed cell migration and invasion assays at 24 h with no significant effect on cell proliferation. As shown in Figure 2A–F, the numbers of migrated and invaded cells were reduced. Besides, the expression of MMP2 and MMP9 was determined by Western blot assay. Compared with the control, the levels of MMP2 and MMP9 were inhibited in Hep3B and Huh-7 cells treated with different concentrations of paeonol (Figure 2G and H). Thus, these findings indicated that cell migration and invasion were suppressed by paeonol in HCC cells.
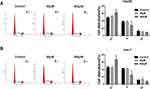
Paeonol down-regulated miR-21-5p level, and silencing of miR-21-5p suppressed cell viability, migration, invasion and promoted apoptosis in Hep3B and Huh-7 cells. To elucidate the relation between paeonol and miR-21-5p, the expression of miR-21-5p in Hep3B and Huh-7 cells with or without paeonol-treatment was detected by qRT-PCR. As exhibited in Figure 3A, the expression level of miR-21-5p was remarkably reduced in paeonol-treated Hep3B and Huh-7 cells compared with the control group. In addition, after transfection with miR-21-5p inhibitor, miR-21-5p expression was significantly decreased in Hep3B and Huh-7 cells (Figure 3B). Furthermore, CCK-8 assay indicated that cell viability was hindered in Hep3B and Huh-7 cells transfected with miR-21-5p inhibitor (Figure 3C). Cell apoptosis was promoted by miR-21-5p inhibitor (Figure 3D). For cell migration and invasion, the number of migrated and invaded cells in both two cell lines transfected with miR-21-5p inhibitor was lower than that in the NC control group (Figure 3E and F). As shown in Figure 3G and H, the expression of Cyclin D1, CDK4, Bcl-2, MMP2 and MMP9 was down-regulated, while Bax level was increased by miR-21-5p knockdown in Hep3B and Huh-7 cells. Collectively, these results suggested that miR-21-5p was down-regulated by paeonol, and its knockdown impeded cell proliferation, migration, invasion and promoted cell apoptosis in Hep3B and Huh-7 cells.
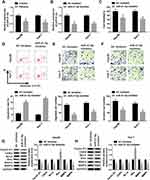
KLF6 Was a Target of miR-21-5p
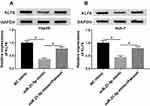
To clarify the underlying molecular mechanism of miR-21-5p in the progression of Hep3B and Huh-7 cells, TargetScan website tool was used to predict the target of miR-21-5p. The binding sites of miR-21-5p on KLF6 3ʹ-UTR and its mutant are exhibited in Figure 4A. Dual-luciferase reporter assay was conducted to confirm the relationship between miR-21-5p and KLF6. After transfection with miR-21-5p mimic and wt-KLF6, the relative luciferase activity was drastically suppressed in wt-KLF6 group, while there was no significant change of luciferase activity in Hep3B and Huh-7 cells co-transfected with miR-21-5p mimic and mut-KLF6 (Figure 4B and C). The efficiency of miR-21-5p transfection was also verified (Figure 4D). In addition, RNA pull-down assay revealed that bio-KLF6 significantly enriched miR-21-5p compared to bio-NC control in Hep3B and Huh-7 cells (Figure 4E). Moreover, Western blot analysis implied that miR-21-5p knockdown obviously elevated the expression of KLF6 in comparison with NC control in Hep3B and Huh-7 cells (Figure 4F and G). In a word, miR-21-5p targeted KLF6 and down-regulated the expression of KLF6.
Knockdown of KLF6 relieved the effect of silenced miR-21-5p on cell viability, migration, invasion and apoptosis in Hep3B and Huh-7 cells. The knockdown efficiency of KLF6 was detected using Western blot assay. The results suggested that KLF6 expression was evidently reduced in Hep3B and Huh-7 cells transfected with si-KLF6 (Figure 5A). Then, Hep3B and Huh-7 cells were introduced with miR-21-5p inhibitor + si-NC or miR-21-5p inhibitor + si-KLF6 to further investigate the interaction between miR-21-5p and KLF6. After transfection with miR-21-5p inhibitor, the expression of KLF6 was markedly enhanced compared with the NC control group, whereas a distinct reduction of KLF6 expression was detected when cells were co-transfected with miR-21-5p inhibitor and si-KLF6 (Figure 5B and C). As shown in Figure 5D–G, inhibition of miR-21-5p reduced cell viability, migration and invasion and elevated cell apoptosis, while the effects were restored after reintroduction with si-KLF6. Furthermore, Western blot analysis suggested that miR-21-5p inhibitor led to a decrease in the levels of Cyclin D1, CDK4, Bcl-2, MMP2 and MMP9, and an increase in Bax expression, which was recovered by KLF6 depletion (Figure 5H and I). These data implied that the down-regulation of KLF6 could reverse the inhibitory effect of miR-21-5p inhibition on HCC progression.
Paeonol Reversed the Suppression of miR-21-5p on KLF6 Expression
To explore the function of miR-21-5p/KLF6/paeonol axis, miR-21-5p were transfected into Hep3B and Huh-7 cells treated with or without paeonol, and the expression of KLF6 was measured by Western blot assay (Figure 6A and B). After transfection with miR-21-5p mimic, the level of KLF6 was conspicuously inhibited in Hep3B and Huh-7 cells, whereas paeonol treatment weakened the suppression on KLF6 expression triggered by miR-21-5p up-regulation. In total, paeonol declined the miR-21-5p-mediated inhibitory effect on KLF6 level in Hep3B and Huh-7 cells.
Paeonol Treatment Impeded the Cell Cycle at the G1 to S Transition
Paeonol treatment inhibited the expression of the cell cycle regulatory proteins (Cyclin D1 and CDK4), so we speculated that paeonol could regulate the cell cycle. Flow cytometry was conducted to detect the cell cycle, and the data indicated that paeonol treatment prominently facilitated cell cycle arrest compared to the control group (Figure 7A and B).
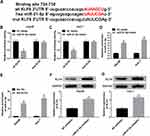
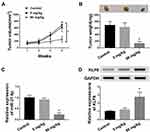
Administration of Paeonol Blocked Tumor Growth in Xenograft Mice
To investigate the role of paeonol in tumorigenesis in vivo, xenograft mice models were established. The results suggested that tumor volume and weight of xenografts treated with 800 μM paeonol were significantly reduced (Figure 8A and B). In addition, paeonol treatment inhibited the expression of miR-21-5p and promoted the expression of KLF6, confirming the results of in vitro experiments (Figure 8C and D). These data certified that paeonol hindered tumor growth in vivo.
Discussion
HCC is one of the common cancers and the third leading cause of death in the world.31 Recent studies revealed that paeonol had been applied in many cancers, including HCC.32 Thus, it is necessary to elucidate the molecular mechanism of paeonol for the treatment of HCC.
In this study, we revealed that paeonol restrained cell viability, migration and invasion, and enhanced cell apoptosis in HCC cells and high concentration of paeonol significantly decreased miR-21-5p expression in HCC cells. Meanwhile, miR-21-5p targeted KLF6 and repressed the expression of KLF6. Also, KLF6 interference relieved the effect of miR-21-5p knockdown on cell viability, apoptosis, migration and invasion in HCC cells. Moreover, the level of KLF6 was apparently inhibited by miR-21-5p overexpression, which was abated by paeonol introduction. These data implied that paeonol modulated HCC progression via regulating miR-21-5p/KLF6 axis.
Paeonol has attracted the attention of researchers all over the world due to its anti-inflammatory, antioxidant and anti-thrombotic effects. The previous study indicated that the proliferation of typhoid bacillus could be inhibited by paeonol.33 Also, through studying the HCC xenograft model, paeonol was identified to reduce oxidative injury and facilitate immunity function in vivo.32 Consistent with the above studies, cell viability, migration and invasion of Hep3B and Huh-7 cells were significantly inhibited, and cell apoptosis was apparently promoted by paeonol in a dose-dependent manner. These results indicated that paeonol might play a crucial role in the progression of HCC.
The regulatory effects of miR-21-5p on tumor development have been extensively identified in many cancers. For instance, miR-21-5p enhanced cell proliferation, migration and invasion in NSCLC via targeting SMAD7.34 Also, miR-21-5p was highly expressed in the recurrence group and might be a potential predictor of gastric cancer recurrence.35 Furthermore, increasing studies determined that miR-21-5p was involved in the progression of HCC. Pu et al reported that miR-21-5p was markedly elevated in the serum of HCC patients.36 Bioinformatics analysis revealed that miR-21-5p might be a novel tumor marker for the diagnosis of HCC.22 However, the network of miR-21-5p in HCC was still unknown. In the current study, we assessed the miR-21-5p expression level in paeonol-treated Hep3B and Huh-7 cells. The level of miR-21-5p was inhibited by paeonol, and its knockdown promoted cell viability, migration and invasion, which was consistent with the previous study.26 Also, miR-21-5p was predicted to target KLF6 mRNA and blocked the expression of KLF6. Hence, we hypothesized that paeonol regulated HCC progression via miR-21-5p/KLF6 axis.
KLF6 is a vital transcription factor and regulates numerous physiology processes, such as cell proliferation, differentiation and adipogenesis. Previous studies showed that KLF6 expression was decreased in HCC,29 and its down-regulation could facilitate tumor activity by targeting the promoter of the oncogene PTTG1.37 To further clarify the function of KLF6 in HCC, we co-transfected si-KLF6 and miR-21-5p into Hep3B and Huh-7 cells. The results indicated that the down-regulation of KLF6 reversed the effects of miR-21-5p silencing on cell viability, apoptosis, migration and invasion. Moreover, paeonol enhanced KLF6 level by restoring the miR-21-5p-mediated effect on KLF6 in Hep3B and Huh-7 cells.
Taken together, this study concluded that paeonol promoted cell viability, migration, invasion and blocked apoptosis by regulating miR-21-5p/KLF6 axis in Hep3B and Huh-7 cells, providing a potential treatment strategy for HCC and new evidence to understand paeonol/miR21-5p/KLF6 axis in HCC. In addition, paeonol hindered tumor growth in vivo.
Disclosure
The authors declare that they have no conflict of interests.
References
1. Bhawna S, Kumar SU. Hepatoprotective activity of some indigenous plants. Int J Pharmtech Res. 2010;1(4):568–572.
2. Kmieć Z. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol. 2001;161:
3. Khemlina G, Ikeda S, Kurzrock R. The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16(1):149. doi:10.1186/s12943-017-0712-x
4. Lee Y, Park H, Lee H, et al. The clinicopathological and prognostic significance of the gross classification of hepatocellular carcinoma. J Pathol Transl Med. 2018;52(2):85–92. doi:10.4132/jptm.2017.11.13
5. Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85. doi:10.1016/j.suronc.2016.03.002
6. Sanyal A, Poklepovic A, Moyneur E, Barghout V. Population-based risk factors and resource utilization for HCC: US perspective. Curr Med Res Opin. 2010;26(9):2183–2191. doi:10.1185/03007995.2010.506375
7. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(10127):1245–1255. doi:10.1016/S0140-6736(18)30010-2
8. Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42(5):1208–1236. doi:10.1002/hep.20933
9. Ozer Etik D, Suna N, Boyacioglu AS. Management of hepatocellular carcinoma: prevention, surveillance, diagnosis, and staging. Exp Clin Transplant. 2017;15(Suppl 2):31–35. doi:10.6002/ect.TOND16.L9
10. Li L, Chen J, Chen X, et al. Serum miRNAs as predictive and preventive biomarker for pre-clinical hepatocellular carcinoma. Cancer Lett. 2016;373(2):234–240. doi:10.1016/j.canlet.2016.01.028
11. Riley CM, Ren TC. Simple method for the determination of paeonol in human and rabbit plasma by high-performance liquid chromatography using solid-phase extraction and ultraviolet detection. J Chromatogr. 1989;489(2):432–437. doi:10.1016/s0378-4347(00)82926-6
12. Ye JM, Deng T, Zhang JB. Influence of paeonol on expression of COX-2 and p27 in HT-29 cells. World J Gastroenterol. 2009;15(35):4410–4414. doi:10.3748/wjg.15.4410
13. Chou TC. Anti-inflammatory and analgesic effects of paeonol in carrageenan-evoked thermal hyperalgesia. Br J Pharmacol. 2003;139(6):1146–1152. doi:10.1038/sj.bjp.0705360
14. Mi XJ, Chen SW, Wang WJ, et al. Anxiolytic-like effect of paeonol in mice. Pharmacol Biochem Behav. 2005;81(3):683–687. doi:10.1016/j.pbb.2005.04.016
15. Zuzic M, Rojo Arias JE, Wohl SG, Busskamp V. Retinal miRNA functions in health and disease. Genes. 2019;10(5):377. doi:10.3390/genes10050377
16. Shirjang S, Mansoori B, Asghari S, et al. MicroRNAs in cancer cell death pathways: apoptosis and necroptosis. Free Radic Biol Med. 2019;139:1–15. doi:10.1016/j.freeradbiomed
17. Subramaniam S, Jeet V, Clements JA, Gunter JH, Batra J. Emergence of MicroRNAs as Key players in cancer cell metabolism. Clin Chem. 2019;65(9):1090–1101. doi:10.1373/clinchem.2018.299651
18. Specjalski K, Jassem E. MicroRNAs: potential biomarkers and targets of therapy in allergic diseases? Arch Immunol Ther Exp. 2019;67(4):213–223. doi:10.1007/s00005-019-00547-4
19. Ma JH, Bu X, Wang JJ, Xie YX. MicroRNA-29-3p regulates hepatocellular carcinoma progression through NF-κB pathway. Clin Lab. 2019;65(5):undefined. doi:10.7754/Clin.Lab.2018.181012
20. Lv L, Wang X, Ma T. microRNA-944 inhibits the malignancy of hepatocellular carcinoma by directly targeting IGF-1R and deactivating the PI3K/Akt signaling pathway. Cancer Manag Res. 2019;11:2531–2543. doi:10.2147/CMAR.S199818
21. Cui Y, Xu HF, Liu MY, et al. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25(15):1890–1898. doi:10.3748/wjg.v25.i15.1890
22. Zhong XZ, Deng Y, Chen G, Yang H. Investigation of the clinical significance and molecular mechanism of miR-21-5p in hepatocellular carcinoma: a systematic review based on 24 studies and bioinformatics investigation. Oncol Lett. 2019;17(1):230–246. doi:10.3892/ol.2018.9627
23. Yoon JS, Kim G, Lee YR, et al. Clinical significance of microRNA-21 expression in disease progression of patients with hepatocellular carcinoma. Biomark Med. 2018;12(10):1105–1114. doi:10.2217/bmm-2018-0096
24. El-Tawdi AH, Matboli M, Shehata HH, et al. Evaluation of circulatory RNA-based biomarker panel in hepatocellular carcinoma. Mol Diagn Ther. 2016;20(3):265–277. doi:10.1007/s40291-016-0200-9
25. Wen Y, Han J, Chen J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137(7):1679–1690. doi:10.1002/ijc.29544
26. Wang J, Chu Y, Xu M, Zhang X, Zhou Y, Xu M. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett. 2019;17(2):2221–2227. doi:10.3892/ol.2018.9843
27. McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381. doi:10.1152/physrev.00058.2009
28. He AD, Xie W, Song W, et al. Platelet releasates promote the proliferation of hepatocellular carcinoma cells by suppressing the expression of KLF6. Sci Rep. 2017;7(1):3989. doi:10.1038/s41598-017-02801-1
29. Wang S, Kang L, Chen X, Zhou H. Frequent down-regulation and deletion of KLF6 in primary hepatocellular carcinoma. J Huazhong Univ Sci Technolog Med Sci. 2010;30(4):470–476. doi:10.1007/s11596-010-0451-3
30. Wu J, Sun C, Wang R, et al. Cardioprotective effect of paeonol against epirubicin-induced heart injury via regulating miR-1 and PI3K/AKT pathway. Chem Biol Interact. 2018;286:17–25. doi:10.1016/j.cbi.2018.02.035
31. Chunhu Z, Suiyu H, Meiqun C, Guilin X, Yunhui L. Antiproliferative and apoptotic effects of paeonol on human hepatocellular carcinoma cells. Anticancer Drugs. 2008;19(4):401–409. doi:10.1097/CAD.0b013e3282f7f4eb
32. Chen B, Ning M, Yang G. Effect of paeonol on antioxidant and immune regulatory activity in hepatocellular carcinoma rats. Molecules. 2012;17(4):4672–4683. doi:10.3390/molecules17044672
33. Li FC, Zhou XL, Mao HL. [A study of paeonol injection on immune functions in rats]. Zhongguo Zhong Xi Yi Jie He Za Zhi. 1994;14(1):37–38. Chinese.
34. Li X, Wu X. MiR-21-5p promotes the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Onco Targets Ther. 2018;11:8445–8454. doi:10.2147/OTT.S172393
35. Park SK, Park YS, Ahn JY, et al. MiR 21-5p as a predictor of recurrence in young gastric cancer patients. J Gastroenterol Hepatol. 2016;31(8):1429–1435. doi:10.1111/jgh.13300
36. Pu C, Huang H, Wang Z, et al. Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front Physiol. 2018;9:930. doi:10.3389/fphys.2018.00930
37. Lee UE, Ghiassi-Nejad Z, Paris AJ, et al. Tumor suppressor activity of KLF6 mediated by downregulation of the PTTG1 oncogene. FEBS Lett. 2010;584(5):1006–1010. doi:10.1016/j.febslet.2010.01.049
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.