Back to Journals » Journal of Inflammation Research » Volume 14
PADI6 Regulates Trophoblast Cell Migration-Invasion Through the Hippo/YAP1 Pathway in Hydatidiform Moles
Authors Huang B , Zhao Y, Zhou L, Gong T, Feng J, Han P, Qian J
Received 30 March 2021
Accepted for publication 29 June 2021
Published 22 July 2021 Volume 2021:14 Pages 3489—3500
DOI https://doi.org/10.2147/JIR.S313422
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Bo Huang,* Yating Zhao,* Lin Zhou,* Tingyu Gong, Jiawen Feng, Peilin Han, Jianhua Qian
Department of Gynecology and Obstetrics, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou City, 310003, Zhejiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianhua Qian
Department of Gynecology and Obstetrics, The First Affiliated Hospital, Zhejiang University School of Medicine, 79 Qingchun Road, Shangcheng District, Hangzhou City, 310003, Zhejiang Province, People’s Republic of China
Tel +8613858028056
Email [email protected]
Purpose: Peptidyl arginine deiminase, type VI (PADI6), a member of the subcortical maternal complex, plays an important role in oocyte growth and the development of fertilized oocytes. Human patients with PADI6 mutations can suffer from multiple reproductive deficiencies including hydatidiform moles and miscarriages. Recent studies have demonstrated that the Hippo signaling pathway plays a central role in the specification of the first cell fates and the maintenance of the human placental trophoblast epithelium. The present study aimed to verify the hypothesis that PADI6 regulates the biological functions of trophoblast cells by targeting YAP1 and to explore the mechanism by which PADI6 accomplishes this in trophoblast cells.
Methods: Villi from HMs and human trophoblast cell lines were used to identify the localization of PADI6 and YAP1 by immunohistochemistry and immunocytochemistry. PADI6 overexpression and knockdown were induced in human trophoblast cells. Co-immunoprecipitation was used to explore the interaction between PADI6 and YAP1. Wound healing, Transwell and EdU staining assays were used to detect migration, invasion and proliferation. Flow cytometric analysis was used to analyze the cell cycle and apoptosis. β-Tubulin and F-actin levels were determined by Western blot, quantitative real-time PCR and phalloidin staining.
Results: The results showed that PADI6 and YAP1 had the same expression pattern in villi and colocalized in the cytotrophoblast. An interaction between PADI6 and YAP1 was also confirmed in human trophoblast cell lines. We found that PADI6 positively regulated the expression of YAP1. Functionally, overexpression of PADI6 promoted cell cycle progression and enhanced migration, invasion, proliferation and apoptosis, whereas downregulation of PADI6 showed the opposite effects.
Conclusion: This study demonstrates that YAP1 is a novel target of PADI6 that serves as an important regulator of trophoblast dysfunction. The crosstalk between the Hippo/YAP1 pathway and the SCMC might be a new topic to explore to uncover the pathological mechanisms of HMs.
Keywords: PADI6, yes-associated protein, trophoblast cell, hydatidiform moles
Introduction
Hydatidiform moles (HMs) are abnormal placentas with variable trophoblastic proliferation and villous hydrops that have a high risk of developing into gestational trophoblastic neoplasia (GTN). Based on their morphological features and underlying genotype, HMs include two varieties, complete hydatidiform moles (CHMs) and partial hydatidiform moles (PHMs). In Western countries and Southeastern Asia, the incidence of HMs is 1 case per 600 pregnancies and 1 case per 60 pregnancies, respectively. A history of HMs (mainly CHMs) is associated with an increased risk of developing GTN, especially choriocarcinoma. Patients with at least two HMs are defined as having recurrent HMs (RHMs), and 1 to 9% of patients with a prior HM have RHMs. To date, it is well established that RHMs are highly related to maternal-effect genes (MEGs). MEGs are a class of genes encoding transcripts that are expressed exclusively in oocytes and early embryos and essential for early embryonic development. As previously reported, mutations in MEGs are associated with HMs. The subcortical maternal complex (SCMC), all components of which are encoded by MEGs, is a multiprotein complex expressed in oocytes and preimplantation embryos. The SCMC regulates a number of essential cellular processes during the oocyte-to-embryo transition, such as spindle assembly, chromosome alignment and symmetric cell division in cleavage-stage embryos. In humans, mutations in SCMC proteins cause various developmental abnormalities, including early embryonic arrest and reproductive failure. For example, mutations in NLRP7 and KHDC3L, two MEGs encoding members of the SCMC, have been shown to account for approximately 55% and 5–10% of RHM cases, respectively. In addition, mutation in TLE6, another member of the SCMC, can result in the earliest known human embryonic lethality phenotype.
Recently, it has been revealed that peptidyl arginine deiminase, type VI (PADI6), one of the MEGs encoding a member of the SCMC, plays an important role in oocyte growth and the development of fertilized oocytes beyond the two-cell stage. Studies have demonstrated that PADI6-deficient mice are infertile due to an early embryo developmental defect, indicating an essential role of PADI6 in female fertility. Previous research also uncovered the involvement of PADI6 to other forms of reproductive deficiency and strengthened the association between PADI6 and infertility, miscarriages, and molar pregnancies. At the cellular level, PADI6 is the first oocyte-specific protein identified to localize to cytoplasmic lattices (CPLs) in the mouse. Cytoskeletal reorganization associated with PADI6 plays a critical role in the regulation of organelle positioning and redistribution. Previous research has found that some patients with PADI6 mutations suffer from multiple reproductive deficiencies, including HMs and miscarriages. Nevertheless, there is still limited evidence identifying the factors involved in the pathogenesis of HM. Trophoblastic proliferation is also a remarkable characteristic, yet it has received little research focus. The underlying mechanisms of HM-associated trophoblast hyperplasia remain largely elusive. Abi Nahed and colleagues reported that NLRP7 protein is directly involved in trophoblast migration, invasion and proliferation in fetal growth restriction. This result suggests that MEGs may also be involved in the dysfunction of trophoblasts in HMs.
YAP1, also known as YAP, is a crucial downstream modulator of the Hippo signaling pathway and plays an increasingly recognized role in the development of cancers by controlling cancer cell growth and invasion in mammals. Existing studies have demonstrated poor YAP1 expression in preeclamptic placentas as a major factor involved in trophoblast dysfunction. The Hippo pathway can be activated by phosphorylation of MST1/2 and LATS1/2. Active LATS1/2 then phosphorylates YAP1. Phosphorylated YAP1 can be precisely recognized by 14-3-3 proteins, leading to subsequent proteasomal degradation or cytoplasmic retention. Interestingly, it was previously reported that 14-3-3 proteins can also bind to PADI6 in a cell cycle- and phosphorylation-dependent manner. Two novel 14-3-3 binding sites have been identified in PADI6, suggesting that there may be an actual role of PADI6 in the regulation of 14-3-3 proteins. Taken together, the data suggest that there may be potential associations between PADI6, 14-3-3 proteins and YAP1, and this association might contribute to the formation of HMs.
However, the role of PADI6 in trophoblast dysfunction in HM and its underlying mechanisms remain unclarified. Therefore, this study was carried out with the aim of identifying the interaction between PADI6 and YAP1 and exploring the possible regulatory effects of this interaction on YAP1 in the progression of HMs.
Materials and Methods
Patients and Controls
Ten formalin-fixed paraffin-embedded tissues of PHMs, CHMs and normal placentas of the first trimester tissues were collected, respectively. Normal placentas tissues were used as control group (NC). All patients were from the First Affiliated Hospital, Zhejiang University School of Medicine. The study was approved by the Institutional Review Board of the First Affiliated Hospital, Zhejiang University School of Medicine (Number: 2020IIT339), and was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent.
Cell Culture
Two human trophoblast cell lines (HTR-8/Svneo and SWAN71) and two human choriocarcinoma cell lines (JAR and JEG3) were used in this study. All cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). HTR-8/Svneo and JAR were maintained in RPMI-1640 medium (Gibco, Grand Island, NY, USA). SWAN71 and JEG3 were maintained in High Glucose-DMEM medium and MEM medium, respectively. All medium supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Gibco) in an incubator at 37°C with 5% CO2.
Cell Transfection
PADI6 small interfering RNA (siRNA) and negative controls were synthesized by SunYa (Shanghai, China), and the sequences are listed in Table 1. Lentiviruses were obtained from Hanbio (Shanghai, China). For siRNA transfection, HTR-8/SVneo and JAR cells were seeded in 6-well plates, and cell transfection was performed when the cells reached 70–90% confluence. siRNA (50 nM) was transfected into cells using Lipofectamine™ 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) for 48 hours. LV-PADI6 or LV-GFP virus was added at a multiplicity of infection of 20, and cells were exposed to virus for 24 hours and selected with 1 μg/mL puromycin for 3 days. The purpose of cell transfection was to conduct functional experiments to elucidate the roles of PADI6 in the biological function of trophoblast cells.
 |
Table 1 siRNA Sequence for PADI6 |
EdU Staining
Cells were seeded in 24-well plates at a density of 2×103 cells/well. EdU (Beyotime, Shanghai, China) was added to the medium to a final concentration of 10 μM at 48 hours post-seeding, followed by 2 hours of incubation at 37°C. After 2 hours, the EdU solution was discarded, and the cells were washed twice with PBS. Then, cells were fixed using methanol solution. Next, a penetrating agent was added and incubated with the cells for 15 min. Subsequently, the cells were incubated with Azide 594 staining solution for 10 min, followed by incubation with DAPI (100 µL) at room temperature for 30 min. The staining result was examined under an Olympus BX51 immunofluorescence microscope (Olympus, Tokyo, Japan).
Transwell Assay
A total of 5×104 cells resuspended in 200 μL serum-free medium were plated in each Transwell chamber precoated with phenol red-free Matrigel. The Transwell chambers were placed in 24-well plates containing 800 μL culture medium supplemented with 20% FBS in each well. After 24 hours of incubation in a 37°C incubator, the cells and the Matrigel on the top surface of the Transwell microporous membrane were wiped off with a cotton swab. The cells on the bottom surface of the membrane were fixed and stained with hematoxylin (Solarbio). Under a 100× inverted microscope, five fields on each membrane were selected, and the numbers of invading cells were counted.
Wound Healing Assay
A wound healing assay was used to evaluate cell migration. HTR-8/SVneo and JAR cells were inoculated into 6-well plates. When the cells reached 90% confluence, a 200 μL pipette tip was used to evenly draw a line at the bottom of the incubator plate. After being washed with PBS, the cells were incubated with serum-free medium for 24 hours. The wound was photographed under a microscope at 0 and 24 hours (4× magnification). ImageJ software was used to quantify the area of the wound to calculate the cell migration rate.
Quantitative Real-Time PCR
Total RNA was extracted from HTR-8/SVneo and JAR cells using TRIzol reagent (Takara Biomedical Technology, Shiga, Japan). cDNA was synthesized using the aHiScript III RT SuperMix for qPCR kit (Vazyme, Nanjing, China). Quantitative real-time PCR (qRT-PCR) analyses were conducted with ChamQTM Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) according to the manufacturer’s protocol. Quantitative real-time PCR was performed on a CFX-96 real-time PCR detection system (Bio-Rad, CA, USA). The primer sequences used in this study are listed in Table 2.
 |
Table 2 Primer Sequence for qRT-PCR |
Western Blot Analysis
Whole-cell proteins were harvested, and the protein concentrations were measured using a BCA kit (Biosharp, Hefei, China). Equal amounts of protein (20 μg) were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (EMD Millipore, MA, USA); subsequently, the membranes were blocked in 5% nonfat dry milk for 2 hours at room temperature. The membranes were incubated with primary antibodies at 4°C overnight, washed with TBST, and incubated with anti-HRP-conjugated Affinipure goat anti-rabbit/mouse lgG(H+L) (Proteintech, Wuhan, China). The signals were detected using an ECL Western blotting kit (Biosharp, Hefei, China). The primary antibodies used included anti-PADI6 (Thermo Fisher, MA, USA), anti-GAPDH (Proteintech, Wuhan, China), anti-YAP1 (Proteintech, Wuhan, China), anti-FLAG (Sigma, MO, USA), anti-beta tubulin (Proteintech, Wuhan, China), anti-14-3-3 (Proteintech, Wuhan, China), anti-MMP2 (Proteintech, Wuhan, China) and anti-MMP9 (Proteintech, Wuhan, China) antibodies.
Flow Cytometric Analysis of the Cell Cycle and Apoptosis
The cell cycle was investigated by a cell cycle staining kit (MultiSciences, Hangzhou, China) according to the manufacturer’s instructions. The cells were stained with propidium iodide (PI) solution after being washed with PBS. Samples were analyzed with a flow cytometer (BD Biosciences, CA, USA). The cell cycle distribution was assessed for further comparisons. Cell apoptosis was investigated with an Annexin V-APC/7-AAD apoptosis kit (MultiSciences, Hangzhou, China) according to the manufacturer’s instructions. The cells were stained with Annexin V-APC and 7-AAD solutions after being washed with PBS. The samples were analyzed with a flow cytometer (BD Biosciences, CA, USA). The relative ratio of apoptotic cells was assessed for further comparisons.
Immunohistochemistry
Paraffin sections of HMs and normal placental tissues underwent dewaxing, hydration and antigen repair. Following blocking with serum, the sections were incubated with anti-PADI6 antibody (1:100 dilution; Bioss, Beijing, China) at 4°C overnight. The slides were washed with PBS and incubated with a secondary antibody at 37°C for 30 min. Then, streptavidin–horseradish peroxidase was applied at 37°C for 30 min, and the slides were stained with DAB solution. Images were obtained using a direct optical microscope. Yellow-brown staining in the cytoplasm was regarded as positive, and the positive stained cells were counted.
Phalloidin Staining
The cytoskeleton was stained with phalloidin from Beyotime (C2205S, Shanghai, China). One microliter of the 200× phalloidin conjugate stock solution was added to 200 µL of PBS with 1% BSA. The cells were incubated in 3–4% formaldehyde in PBS at room temperature for 20 min. Fixed cells were washed with PBS 3 times. Then, 100 µL of 1× phalloidin conjugate working solution was added to each well and incubated at room temperature for 60 min. The cells were mounted using mounting buffer containing DAPI, and cytoskeletal staining was observed using a fluorescence microscope.
Cell Immunofluorescence
Cells were washed three times with cold PBS, fixed with 4% paraformaldehyde for 30 min at room temperature, permeabilized in 0.2% Triton X-100 for 20 min at room temperature, and then incubated with 1% BSA for 1 hour at room temperature. Next, they were stained with primary anti-PADI6 (Bioss, Beijing, China) and anti-YAP1 (Proteintech, Wuhan, China) antibodies overnight at 4°C, and secondary CoraLite594-conjugated goat anti-mouse IgG (H+L) (Proteintech, Wuhan, China) and CoraLite488-conjugated goat anti-rabbit IgG (H+L) (Proteintech, Wuhan, China) antibodies were applied for 1 hour at room temperature. Cell nuclei were stained with DAPI (Biosharp, Hefei, China). Fluorescence was examined under a Leica STELLARIS 8 confocal microscope (Leica, Wetzlar, Germany).
Co-Immunoprecipitation
For in vitro co-immunoprecipitation (co-IP) analysis, FLAG-tagged proteins were produced in HTR-8/Svneo and JAR cells transfected with LV-PADI6. Total cell lysates were incubated overnight at 4°C with anti-YAP1 antibodies (Proteintech, Wuhan, China), anti-FLAG antibodies (Sigma, MO, USA), or normal IgG (Beyotime, Shanghai, China) as a control. Antibody-antigen complexes were precleared with Protein A/G PLUS-Agarose (Beyotime, Shanghai, China). After several washes, samples were boiled and analyzed by immunoblot.
Statistical Analyses
All experimental data were processed by SPSS 21.0 statistical software (IBMCorp., Armonk, NY, USA). Measurement data are expressed as the mean ± standard deviation of three independent tests. Comparisons between two groups with normal distribution and homogeneity of variance were performed with paired t-tests. Comparisons between two groups with normal distribution and homogeneity of variance were performed with unpaired t-tests. Data comparisons between multiple groups were performed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. The difference was statistically significant at p<0.05.
Results
The Expression Patterns of PADI6 and YAP1
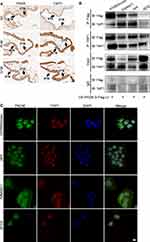
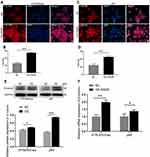
In the present study, we detected the expression of PADI6 and YAP1 in villi tissues from HM patients and normal placentas. The results showed that PADI6 and YAP1 were located in the nucleus of cytotrophoblast cells, and a coexpression pattern was identified (Figure 1A). In addition, immunofluorescence was performed to observe the location of PADI6 and YAP1 in human trophoblast cell lines, and the results suggested that PADI6 and YAP1 colocalize within nuclei in HTR-8/SVneo, Swan71, JAR and JEG3 cells (Figure 1C). Subsequently, to further confirm the interaction of PADI6 and YAP1, an oe-PADI6 lentivirus vector was transfected into four types of trophoblast cell lines, HTR-8/SVneo, Swan71, JAR and JEG3 cells, and we observed via co-IP assays that PADI6 interacted with YAP1 in all four trophoblast cell lines (Figure 1B).
Overexpression of PADI6 Upregulated the Expression of YAP1
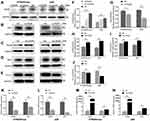
In an attempt to elucidate the association of PADI6 with YAP1 in trophoblast cells, the normal human trophoblast cell line HTR-8/SVneo and the choriocarcinoma cell line JAR were transfected with si-PADI6 (knock down) and oe-PADI6 (overexpression) lentivirus vectors. The transfection efficiency was assessed by qRT-PCR and Western blot, which demonstrated that PADI6 expression was markedly decreased following transfection of si-PADI6 compared with the level in cells transfected with si-NC, and the expression of YAP1 was inhibited by the decrease in PADI6 (Figure 2A, B, F, G, K and L). In HTR-8/SVneo and JAR cells transfected with LV-PADI6-flag, a significant increase in PADI6 was observed, accompanied by upregulation of YAP1 (Figure 2C, D, H, I, M and N). 14-3-3 proteins, regulators of YAP1, were also notably decreased at the protein level in trophoblast cells after transfection with oe-PADI6 compared with the levels in controls (Figure 2E and J). These results suggest that PADI6 can positively regulate the expression of YAP1 through 14-3-3 proteins.
Overexpression of PADI6 Promoted the Migration and Invasion of HTR-8/SVneo and JAR Cells
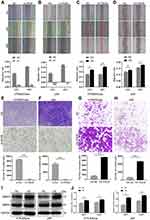
To determine the role of PADI6 in the biological function of trophoblast cells, PADI6 knockdown and overexpression were induced in both HTR-8/SVneo and JAR cells. Then, we performed a wound healing assay and revealed that PADI6 promoted the migration of HTR-8/SVneo and JAR cells (Figure 3A–D). In addition, Transwell assays showed that the numbers of migrated and invaded HTR-8/SVneo and JAR cells were increased by PADI6 upregulation (Figure 3E–H). Western blot analysis showed that transfection of oe-PADI6 led to increased MMP2 and MMP9 protein expression (Figure 3I and J). Together, these results indicate that PADI6 overexpression in trophoblast cells promotes migration and invasion by upregulating MMP2 and MMP9.
Overexpression of PADI6 Enhanced the Proliferation and Apoptosis of HTR-8/SVneo and JAR Cells
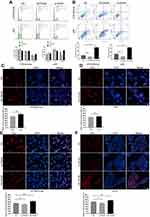
Next, we attempted to elucidate the regulatory effect of PADI6 on trophoblast cell proliferation and apoptosis. HTR-8/SVneo and JAR cells were transfected with si-PADI6 or oe-PADI6 lentivirus vectors. Following transfection, cell cycle distribution, cell proliferation and apoptosis were evaluated by flow cytometry and EdU staining. In both cell lines, transfection with si-PADI6 resulted in an increase in the proportion of cells in G0/G1 phase and a decrease in the proportion of cells in G2/M phase (Figure 4A), accompanied by a reduced rate of cell apoptosis (Figure 4B), but the opposite trends were observed in cells transfected with the oe-PADI6 lentivirus vector. The EdU results indicated that upregulation of PADI6 increased the proliferation rate of HTR-8/SVneo and JAR cells (Figure 4C and D). HTR and JEG3 cells in invalidated and overexpressing conditions of PADI6 were used to verify this results, and obtained consistent outcome (Figure 4E and F). Taken together, these data suggest that PADI6 upregulation promotes cell cycle progression and enhances the proliferation and apoptosis of HTR-8/SVneo and JAR cells.
Overexpression of PADI6 Regulated Cytoskeleton Dynamics
The above experimental results demonstrated that PADI6 overexpression upregulated the expression of YAP1, enhanced the migration and invasion of HTR-8/SVneo and JAR cells, and promoted proliferation and the cell cycle process. To further evaluate the role of PADI6 and YAP1 in the biological functions of trophoblast cells, an oe-PADI6 lentivirus vector was transfected into HTR-8/SVneo and JAR cells. F-actin and β-tubulin expression was measured using phalloidin staining and Western blot analysis, respectively. The results showed that the fluorescence intensity of F-actin was significantly increased in oe-PADI6 cell lines (Figure 5A–D). The Western blot analysis indicated that β-tubulin was also significantly increased at both the mRNA and protein levels with overexpression of PADI6 (Figure 5E and F). Taken together, these data suggest that PADI6 effects cytoskeletal dynamics by manipulating F-actin and β-tubulin.
Discussion
As a shuttling protein between the nucleus and cytoplasm, YAP1 mediates cell functions such as invasion, migration and angiogenesis. Dephosphorylated YAP1 translocates into the nucleus, where it interacts with transcription factors and participates in pathophysiological processes. After binding 14-3-3 proteins, phosphorylation of YAP1 results in cytoplasmic retention and functional inactivation, leading to subsequent proteasomal degradation. Increasing evidence indicates that the Hippo-YAP1 pathway participates in early embryo development. YAP1 plays a pivotal role in the maintenance of the human placental trophoblast epithelium. Biswarup et al also demonstrated that TEAD4, an effector of the Hippo signaling pathway, is essential for the establishment of pregnancy in postimplantation embryos. In the present study, we reported for the first time that overexpression of PADI6 induced upregulation of YAP1 in trophoblast cell lines. Importantly, our results showed that PADI6 and YAP1 colocalized in the nuclei of trophoblast cells, and co-IP further confirmed that they interacted with each other as a protein complex. In vitro experiments demonstrated that PADI6 overexpression competed with YAP1 for binding with 14-3-3 proteins, leading to a decrease in the cytoplasmic retention of YAP1. Since PADI6 and YAP1 both participate in early embryo development, the interaction between PADI6 and YAP1 might indicate that they are not only physically bound but also functionally linked.
It has been reported that repression of YAP1 can help to impair trophoblast invasion and migration and suppress cell apoptosis. YAP1 expression has also been found to be primarily associated with trophoblast stemness and proliferation. Our results showed that PADI6 participated in multiple biological functions of trophoblast cells, including migration, invasion, proliferation and apoptosis. In vitro experiments confirmed that PADI6 induces excessive expression of YAP1 at both the mRNA and protein levels in trophoblast cells, and increased expression of MMP2 and MMP9 was also observed after PADI6 overexpression, indicating that the effect of PADI6 on trophoblast cells might be carried out through YAP1. Previous studies found that PADI6 plays a critical role in microtubule-mediated organelle positioning and movement during oocyte maturation, but its effect on trophoblast cells has not been fully uncovered. Emerging evidence suggests that the actin cytoskeleton, the intermediate filament network, and microtubules are involved in the regulation of cell motility. In the present study, a remarkable increase in F-actin and β-tubulin was also observed in trophoblast cells following transfection with oe-PADI6. It is likely that the changes in F-actin and β-tubulin influenced cytoskeletal dynamics, subsequently leading to altered cell migration. Another member of the SCMC, NLRP7, was also found to be involved in trophoblast proliferation. Taken together, our findings support the observation that members of the SCMC, such as PADI6 and NLRP7, participate in the biological functions of trophoblast cells. The key findings of our study further confirm that the proliferation-promoting and metastasis-promoting roles of PADI6 in trophoblast cells might be mediated via upregulation of YAP1.
In conclusion, these results suggest that PADI6 enhances the migration, invasion, proliferation and apoptosis of trophoblast cells by targeting YAP1. PADI6 is proposed to interact with YAP1 in HM processes. However, the data of this study were obtained from cell lines, and further studies are needed to elucidate the underlying mechanism by which PADI6 targets YAP1 in vivo. Nevertheless, PADI6 could participate in the regulation of the Hippo/YAP1 pathway, this interaction probably indicates crosstalk between MEGs and the Hippo/YAP1 pathway, which may reveal a new avenue for acquiring a full understanding of the pathogenesis of HMs.
Abbreviations
HM, Hydatidiform moles; CHM, complete hydatidiform mole; PHM, partial hydatidiform mole; MEGs, maternal-effect genes; SCMC, subcortical maternal complex; OE, overexpression; STB, syncytiotrophoblast; CTB, cytotrophoblast.
Funding
This work was supported by National Natural Science Foundation of China (82071665 and 81902269), the key Research and Development Program of Zhejiang Province, China (2020C03116) and Center for Uterine Cancer Diagnosis &Therapy Research of Zhejiang Province, China.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Wang Q, Fu J, Hu L, et al. Prophylactic chemotherapy for hydatidiform mole to prevent gestational trophoblastic neoplasia. Cochrane Database Syst Rev. 2017;9:CD007289. doi:10.1002/14651858.CD007289.pub3
2. Rezaei M, Suresh B, Bereke E, et al. Novel pathogenic variants in NLRP7, NLRP5, and PADI6 in patients with recurrent hydatidiform moles and reproductive failure. Clin Genet. 2021;99(6):823–828. doi:10.1111/cge.13941
3. Kaneki E, Kobayashi H, Hirakawa T, Matsuda T, Kato H, Wake N. Incidence of postmolar gestational trophoblastic disease in androgenetic moles and the morphological features associated with low risk postmolar gestational trophoblastic disease. Cancer Sci. 2010;101(7):1717–1721. doi:10.1111/j.1349-7006.2010.01602.x
4. Kan M, Yamamoto E, Niimi K, et al. Gestational trophoblastic neoplasia and pregnancy outcome after routine second curettage for hydatidiform mole a Retrospective Observational Study. J Reprod Med. 2016;61(7–8):373–379.
5. Bebbere D, Masala L, Albertini DF, Ledda S. The subcortical maternal complex: multiple functions for one biological structure? J Assist Reprod Genet. 2016;33(11):1431–1438. doi:10.1007/s10815-016-0788-z
6. Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15(3):416–425. doi:10.1016/j.devcel.2008.07.010
7. Lu X, Gao Z, Qin D, Li L. A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol Med. 2017;23(11):1014–1023. doi:10.1016/j.molmed.2017.09.004
8. Alazami AM, Awad SM, Coskun S, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16(1):240. doi:10.1186/s13059-015-0792-0
9. Docherty LE, Rezwan FI, Poole RL, et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun. 2015;6(1):8086. doi:10.1038/ncomms9086
10. Murdoch S, Djuric U, Mazhar B, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38(3):300–302. doi:10.1038/ng1740
11. Parry DA, Logan CV, Hayward BE, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89(3):451–458. doi:10.1016/j.ajhg.2011.08.002
12. Maddirevula S, Coskun S, Awartani K, Alsaif H, Abdulwahab FM, Alkuraya FS. The human knockout phenotype of PADI6 is female sterility caused by cleavage failure of their fertilized eggs. Clin Genet. 2017;91(2):344–345. doi:10.1111/cge.12866
13. Xu Y, Shi Y, Fu J, et al. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99(3):744–752. doi:10.1016/j.ajhg.2016.06.024
14. Qian J, Nguyen NMP, Rezaei M, et al. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet. 2018;26(7):1007–1013. doi:10.1038/s41431-018-0141-3
15. Wright PW, Bolling LC, Calvert ME, et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003;256(1):73–88. doi:10.1016/s0012-1606(02)00126-4
16. Esposito G, Vitale AM, Leijten FP, et al. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273(1–2):25–31. doi:10.1016/j.mce.2007.05.005
17. Shaaban AM, Rezvani M, Haroun RR, et al. Gestational trophoblastic disease: clinical and imaging features. Radiographics. 2017;37(2):681–700. doi:10.1148/rg.2017160140
18. Abi Nahed R, Reynaud D, Borg AJ, et al. NLRP7 is increased in human idiopathic fetal growth restriction and plays a critical role in trophoblast differentiation. J Mol Med. 2019;97(3):355–367. doi:10.1007/s00109-018-01737-x
19. Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–79. doi:10.1038/nrc3876
20. Ling HH, Kuo CC, Lin BX, Huang YH, Lin CW. Elevation of YAP promotes the epithelial-mesenchymal transition and tumor aggressiveness in colorectal cancer. Exp Cell Res. 2017;350(1):218–225. doi:10.1016/j.yexcr.2016.11.024
21. Sun M, Na Q, Huang L, et al. YAP is decreased in preeclampsia and regulates invasion and apoptosis of HTR-8/SVneo. Reprod Sci. 2018;25(9):1382–1393. doi:10.1177/1933719117746784
22. Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154(6):1342–1355. doi:10.1016/j.cell.2013.08.025
23. Ni L, Zheng Y, Hara M, Pan D, Luo X. Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 2015;29(13):1416–1431. doi:10.1101/gad.264929.115
24. Bae JS, Kim SM, Lee H. The Hippo signaling pathway provides novel anti-cancer drug targets. Oncotarget. 2017;8(9):16084–16098. doi:10.18632/oncotarget.14306
25. Sharma A, Yerra VG, Kumar A. Emerging role of Hippo signalling in pancreatic biology: YAP re-expression and plausible link to islet cell apoptosis and replication. Biochimie. 2017;133:56–65. doi:10.1016/j.biochi.2016.12.009
26. Liu CY, Zha ZY, Zhou X, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285(48):37159–37169. doi:10.1074/jbc.M110.152942
27. Rose R, Rose M, Ottmann C. Identification and structural characterization of two 14-3-3 binding sites in the human peptidylarginine deiminase type VI. J Struct Biol. 2012;180(1):65–72. doi:10.1016/j.jsb.2012.05.010
28. Wang H, Xu P, Luo X, et al. Phosphorylation of yes-associated protein impairs trophoblast invasion and migration: implications for the pathogenesis of fetal growth restrictiondagger. Biol Reprod. 2020;103(4):866–879. doi:10.1093/biolre/ioaa112
29. Sasaki H. Roles and regulations of Hippo signaling during preimplantation mouse development. Dev Growth Differ. 2017;59(1):12–20. doi:10.1111/dgd.12335
30. Meinhardt G, Haider S, Kunihs V, et al. Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc Natl Acad Sci U S A. 2020;117(24):13562–13570. doi:10.1073/pnas.2002630117
31. Saha B, Ganguly A. TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: an implication in early human pregnancy loss. Proc Natl Acad Sci U S A. 2020;117(30):17864–17875. doi:10.1073/pnas.2002449117
32. Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. doi:10.1126/science.1175862
33. Tang DD, Gerlach BD. The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir Res. 2017;18(1):54. doi:10.1186/s12931-017-0544-7
34. Wang H, Yao H, Yi B, et al. MicroRNA-638 inhibits human airway smooth muscle cell proliferation and migration through targeting cyclin D1 and NOR1. J Cell Physiol. 2018;234(1):369–381. doi:10.1002/jcp.26930
35. Garcin C, Straube A, Malliri A, Caswell P, Ballestrem C, Hurlstone A. Microtubules in cell migration. Essays Biochem. 2019;63(5):509–520. doi:10.1042/EBC20190016
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.