Back to Journals » OncoTargets and Therapy » Volume 13
Paclitaxel Suppresses Hepatocellular Carcinoma Tumorigenesis Through Regulating Circ-BIRC6/miR-877-5p/YWHAZ Axis
Authors Liu Y, Guo J, Shen K, Wang R, Chen C, Liao Z, Zhou J
Received 11 May 2020
Accepted for publication 19 August 2020
Published 22 September 2020 Volume 2020:13 Pages 9377—9388
DOI https://doi.org/10.2147/OTT.S261700
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Yi Liu,1 Jianchao Guo,2 Ka Shen,3 Renlong Wang,3 Cheng Chen,3 Zhiyuan Liao,3 Jianbo Zhou3
1Department of Traditional Chinese Medicine, Suizhou Hospital, Hubei University of Medicine, Suizhou 441300, People’s Republic of China; 2Department of Gastroenterology, Suizhou Hospital, Hubei University of Medicine, Suizhou 441300, People’s Republic of China; 3Department of General Surgery, Suizhou Hospital, Hubei University of Medicine, Suizhou 441300, People’s Republic of China
Correspondence: Jianbo Zhou Tel +86-139722288169
Email [email protected]
Background: Paclitaxel is an effective chemotherapeutic agent for the treatment of cancer patients. Accumulating evidence suggests that circular RNAs (circRNAs) play critical roles in the occurrence and development of human cancers. However, there are few studies on interactions between paclitaxel and circRNAs in hepatocellular carcinoma (HCC).
Materials and Methods: Cell counting kit-8 (CCK-8) assay and colony formation assay were conducted to determine cell proliferation. Cell apoptosis was assessed by flow cytometry. The expression levels of circRNA baculoviral IAP repeat-containing 6 (circ-BIRC6), microRNA-877-5p (miR-877-5p), and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (YWHAZ) were detected by quantitative real-time polymerase chain reaction (qRT-PCR). The mice xenograft model was established to investigate the roles of circ-BIRC6 and paclitaxel in vivo. The interaction between miR-877-5p and circ-BIRC6 or YWHAZ was predicted by bioinformatics analysis and verified by dual-luciferase reporter assay. Western blot assay was applied for measuring the protein expression of YWHAZ.
Results: Paclitaxel suppressed HCC tumorigenesis through decreasing cell proliferation and accelerating apoptosis. Circ-BIRC6 and YWHAZ were upregulated, and miR-877-5p was downregulated in HCC tissues and cells. Paclitaxel treatment inhibited the expression of circ-BIRC6 and YWHAZ while promoted the expression of miR-877-5p. Circ-BIRC6 overexpression or miR-877-5p interference reversed the inhibitory effect of paclitaxel on HCC tumorigenesis. Moreover, miR-877-5p could specially bind to YWHAZ, and its knockdown abated the suppressive effect of circ-BIRC6 depletion on HCC tumorigenesis. Additionally, YWHAZ was identified as a direct target of miR-877-5p. Besides, circ-BIRC6 functioned as a molecular sponge of miR-877-5p to regulate YWHAZ expression.
Conclusion: Paclitaxel limited HCC tumorigenesis via modulating circ-BIRC6/miR-877-5p/YWHAZ axis, providing a novel therapeutic approach for the treatment of HCC.
Keywords: hepatocellular carcinoma, paclitaxel, circ-BIRC6, miR-877-5p, YWHAZ
Introduction
Liver cancer is one of the most lethal and prevalent cancers, causing approximately 782, 000 deaths in 2018 worldwide.1 Hepatocellular carcinoma (HCC) accounts for 85–90% of primary liver cancer.2 In recent years, despite new advances in the development of treatment strategies, the prognosis of patients remains poor as most HCC patients are usually diagnosed at advanced stages.3 Therefore, it is important to explain the underlying mechanisms of HCC tumorigenesis and improve the therapeutic effect and prognosis of HCC patients.
The systemic chemotherapy or combination chemotherapy of paclitaxel is applied for treating multiple types of cancer, including HCC.4,5 The effective antitumor effect of paclitaxel is mediated by a variety of mechanisms. Paclitaxel directly induces DNA damage and apoptosis, leading to cell death.6 Understanding the underlying molecular mechanisms of paclitaxel-mediated cell death can help improve therapeutic efficacy and select the appropriate combinations with other therapies.
Circular RNAs (circRNA), a new type of endogenous non-coding (nc) RNAs, covalently connect the 5ʹ and 3ʹ ends of the RNA to create closed-loop structures.7 It has been reported that circRNAs play critical roles in many biological processes and have higher stability and conservation than linear RNA.8 Up to now, some studies have shown that circRNAs have essential roles in the tumorigenesis of multiple malignancies, such as cervical,9 bladder,10 breast,11 and pancreatic12 cancers. CircRNA baculoviral IAP repeat-containing 6 (circ-BIRC6; ID: hsa_circ_0003288, chr2:32,703,702–32,718,734) has been found to act as an oncogene in HCC.13 However, more functions of circ-BIRC6 and its association with paclitaxel in HCC are still largely unknown.
It is well known that circRNAs contribute to cancer development through serving as microRNA (miRNA) sponges to modulate gene and protein expression.14 MiRNAs, a kind of small RNA (about 22 nucleotides), lack protein-coding capacity and inversely modulate gene expression through binding to complementary sequences on target mRNA.15,16 MiR-877-5p has been suggested to act as an anti-oncogene in many cancers, including HCC.17 Some studies have demonstrated that miRNAs are involved in paclitaxel-mediated function in multiple cancers.18 However, the interactions among miR-877-5p, circ-BIRC6, and paclitaxel have not been clarified. Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (YWHAZ, also known as 14-3-3ζ), has been observed to be frequently upregulated in HCC.19 Bioinformatics tool exhibits the putative binding sites between miR-877-5p and circ-BIRC6 or YWHAZ, implying that circ-BIRC6 may function as a molecular sponge for miR-877-5p to regulate YWHAZ expression.
In this work, we explored the effects of paclitaxel and circ-BIRC6 on HCC cell proliferation and apoptosis. Moreover, we investigated whether the functions of paclitaxel were regulated by circ-BIRC6 and miR-877-5p. Additionally, the regulatory network of circ-BIRC6/miR-877-5p/YWHAZ axis was explored in HCC cells. The purpose of our research was to provide a promising avenue for treating HCC.
Materials and Methods
Patient Samples
In our research, HCC tissues (n=30) and adjacent non-tumor tissues (n=30) were obtained from patients who underwent surgery at Suizhou Hospital, Hubei University of Medicine. None of the patients received any treatment before surgery. These tissue samples were timely frozen in liquid nitrogen until use. These patients had provided informed consent, and this procedure was granted by the Ethics Committee of Suizhou Hospital, Hubei University of Medicine (with the approval No. 20,190,413), which was in accordance with the Declaration of Helsinki Principles. Informed consent was obtained from all participants.
Cell Culture and Transfection
Human HCC cells (Huh-7 and Hep3B) and human normal liver cells (THLE-2) were purchased from COBIOER (Nanjing, China). These cells were cultured in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (Gibco, Carlsbad, CA, USA) and antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin) at 37°C in a humidified atmosphere with 5% CO2. Paclitaxel (Sigma-Aldrich, St. Louis, MO, USA) was used to treat Huh-7 and Hep3B cells.
Circ-BIRC6 overexpression plasmid (circ-BIRC6) and its control (pcDNA), small interfering RNA against circ-BIRC6 (si-circ-BIRC6) and its control (si-NC), mimic or inhibitor of miR-877-5p (miR-877-5p or anti-miR-877-5p) and their controls (miR-NC or anti-miR-NC) were provided by GenePharma (Jiangsu, China). Lentivirus-mediated short hairpin RNA interference targeting circ-BIRC6 (sh-circ-BIRC6) and matched control (sh-NC) were bought from Genechem (Shanghai, China). According to the manufacturer’s suggested protocol, transient transfection was performed in our research by using Lipofectamine 3000 Reagent (Invitrogen).
Cell Viability Assay
Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China) was applied for detecting cell viability. In brief, Huh-7 and Hep3B cells were seeded in 96-well plates one day prior to transfection, and different concentrations of paclitaxel (2, 10, and 20 nM) were added after transfection. CCK-8 (10 μL) solution was added to per well and incubated for an additional 3 h. The microplate reader (Bio-Rad, Hercules, CA, USA) at 450 nm was employed to measure the optical density (OD) value at 48 h.
Colony Formation Assay
After treatment or/and transfection, Huh-7 and Hep3B cells were counted and placed in 6-well plates. The culture medium was updated every 3 days during colony growth. After incubation for 2 weeks, the cells were washed twice using the phosphate-buffered saline (PBS), followed by fixing with paraformaldehyde (4%) and staining with crystal violet (0.1%, Sigma-Aldrich). The number of colonies was calculated only if they contained more than 50 cells.
Cell Apoptosis Assay
After treatment or/and transfection, Huh-7 and Hep3B cells were harvested, centrifuged, and re-suspended, followed by staining with Annexin V-fluorescein isothiocyanate (FITC)/Propidium Iodide (PI) (Key GEN BioTECH, Jiangsu, China). Flow cytometry (Partec AG, Arlesheim, Switzerland) was used to analyze apoptosis.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Trizol reagent (Invitrogen) was utilized to gain total RNA from tissue samples and cells. The first strand of complementary DNA (cDNA) was synthesized with the FastQuant RT kit (Tiangen Biotech, Beijing, China). The qRT-PCR was carried out using SYBR Select Master Mix (Tiangen Biotech) on an ABI 7300 system (Thermo Fisher Scientific, Waltham, MA, USA). The primers used for qRT-PCR were exhibited as below: circ-BIRC6 (Forward, 5ʹ-TGAAAGGTTCTTGCACGCAT-3ʹ; Reverse, 5ʹ-GCTGGGGTTCGTTCACAATC-3ʹ); miR-877-5p (Forward, 5ʹ-GTAGAGGAGATGGCGCAGGG-3ʹ; Reverse, 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ); YWHAZ (Forward, 5ʹ-GTCGATCAGTCACAACAAGCA-3ʹ; Reverse, 5ʹ-CCGATGTCCACAATGTCAAGT-3ʹ); glyceraldehyde3-phosphate dehydrogenase (GAPDH) (Forward, 5ʹ-ACCACAGTCCATGCCATCAC-3ʹ; Reverse, 5ʹ-TCCACCACCCTGTTGCTGTA-3ʹ); U6 (Forward, 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ; Reverse, 5ʹ-ACGCTTCACGAATTTGCGTGTC-3ʹ). The expression levels of circ-BIRC6, YWHAZ and miR-877-5p were evaluated using the 2−ΔΔCt method and normalized with GAPDH (for circ-BIRC6 and YWHAZ) and U6 (for miR-877-5p).
Tumor Xenograft Model
Animal experiments have been approved by the Animal Care and Use Committee of Suizhou Hospital, Hubei University of Medicine, and performed according to the guidelines for laboratory animal welfare (GB T 35,892–2018). Five-week-old male BALB/c nude mice (n=6/group) were bought from Huafukang (Beijing, China). They were raised in an SPF environment, with 12 h light/dark cycles, and constant and suitable temperature (25°C) and humidity (60%). All mice were allowed free access to drinking water and sterilized standard diet. We made reasonable efforts to minimize the suffering of animals. The sh-circ-BIRC6 or sh-NC was transfected into Hep3B cells. Subsequently, stably transfected cells (4×106/0.2 mL PBS) were subcutaneously injected in BALB/c nude mice. Transfected mice were divided into 4 groups: sh-NC + PBS, sh-circ-BIRC6 + PBS, sh-NC + paclitaxel, sh-circ-BIRC6 + paclitaxel. From the 7th day, mice were treated with paclitaxel (30 mg/kg) every 3 days for 5 cycles. The tumor size was tested using a caliper every 3 days and calculated using the formula: 0.5 ×length × width2. After injection for 22 days, all mice would be sacrificed by cervical dislocation after deep anesthesia with 2% isoflurane (Baxter Healthcare Corporation, Deerfield, IL, USA), and the excised tumor tissues were collected for weight and subsequent qRT-PCR analysis.
Dual-Luciferase Reporter Assay
Targeting the relationship between miR-877-5p and circ-BIRC6 or YWHAZ was predicted using the starBase v2.0. Fragments from circ-BIRC6 and YWHAZ 3ʹUTR that contained wild-type (WT) or mutant (MUT) miR-877-5p binding sites were amplified and cloned into pmirGlO luciferase reporter vector (Promega, Madison, WI, USA), named as WT-circ-BIRC6, YWHAZ 3ʹUTR-WT, MUT-circ-BIRC6, and YWHAZ 3ʹUTR-MUT. The reporter plasmids were transfected into Huh-7 and Hep3B cells along with miR-877-5p or miR-NC for 48 h. Finally, a dual-luciferase assay system (Promega) was utilized for measuring the luciferase activity, followed by normalizing to Renilla luciferase activity.
Western Blot Assay
Total protein was extracted using RIPA lysis buffer that contained 1 nM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich). BCA Protein Assay kit (Applygen, Beijing, China) was applied to quantify the protein concentration. Next, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was applied to separate the equal amounts of protein and then polyvinylidene difluoride membranes (Beyotime) were used to transfer the protein. Membranes were blocked using the 5% non-fat milk (Sangon Biotech, Shanghai, China) and then incubated overnight with the primary antibody against YWHAZ (1:500, ab51129, Abcam, Cambridge, MA, USA) or GAPDH (1:2000, ab37168, Abcam), followed by incubation with secondary antibody (1:4000, ab205718, Abcam). After the membranes were completely washed, the enhanced chemiluminescence detection reagent (Tanon, Shanghai, China) was applied to visualize the protein bands. Signal intensity was quantified by ImageJ software, followed by normalizing to GAPDH expression.
Statistical Analysis
In this study, all data from at least three independent experiments were expressed as mean ± standard deviation (SD) and analyzed with Graphpad Prism version 6.0 software (GraphPad Software, San Diego California, USA). The Student’s t-test was applied to compare the significance of differences between two groups, and one-way analysis of variance (ANOVA) was used for multiple-group comparisons. Correlation between miR-877-5p and circ-BIRC6 or YWHAZ was detected by Spearman’s rank correlation. P < 0.05 was considered statistically significant.
Results
Paclitaxel Could Suppress HCC Tumorigenesis by Reducing Cell Proliferation and Promoting Apoptosis
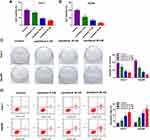
To explore the effect of paclitaxel on HCC tumorigenesis, functional experiments were performed. CCK-8 analysis revealed that cell viability was dose-dependently inhibited in Huh-7 and Hep3B cells after treatment with paclitaxel, and IC50 values of paclitaxel were 8.4 and 3.7 nM in Huh-7 and Hep3B cells, respectively (Figure 1A and B, Supplementary Figure 1A and 1B). Likewise, colony formation assay displayed that the number of colonies was dose-dependently decreased in Huh-7 and Hep3B cells following exposure to paclitaxel (Figure 1C). And the original figures of the colony formation were presented in Supplementary Figures 2–9. Flow cytometry analysis was applied to measure the proportion of apoptotic cells. As presented in Figure 1D, paclitaxel treatment increased cell apoptosis in a dose-dependent manner. These results indicated that paclitaxel inhibited cell proliferation and induced apoptosis in HCC cells.
The Expression Level of Circ-BIRC6 Was Upregulated, and miR-877-5p Was Downregulated in HCC
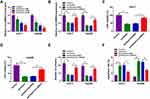
To investigate the potential roles of circ-BIRC6 and miR-877-5p in HCC, their levels were examined by qRT-PCR. As illustrated in Figure 2A and B, the expression of circ-BIRC6 was enhanced in HCC tumor (T) tissues and cells compared with their matched normal (N) tissues. However, the level of miR-877-5p was lowly expressed in HCC tissues and cells in contrast to their corresponding controls (Figure 2C and D). In addition, the correlation between miR-877-5pand circ-BIRC6expression was analyzed in HCC tissues. As presented in Figure 2E, miR-877-5pexpression was negatively correlated with the circ-BIRC6 level in HCC tissues (r=−0.5798, p<0.001). These data implied that circ-BIRC6 and miR-877-5p might play critical roles in HCC tumorigenesis.
Paclitaxel Inhibited HCC Tumorigenesis Through Down-Regulating Circ-BIRC6
To explore the effect of paclitaxel on circ-BIRC6 expression, qRT-PCR assay was carried out. The results showed that paclitaxel treatment dose-dependently reduced the expression of circ-BIRC6 in Huh-7 and Hep3B cells (Figure 3A). Subsequently, the transfection of circ-BIRC6 abated the repressive impact of paclitaxel on the expression of circ-BIRC6 (Figure 3B). Next, we explored whether the function of paclitaxel in HCC tumorigenesis is mediated by circ-BIRC6. We uncovered that overexpression of circ-BIRC6 reversed the inhibitory effect of paclitaxel on proliferation by increasing cell viability and colonies (Figure 3C-E). Moreover, the upregulation of circ-BIRC6 weakened paclitaxel-induced apoptosis in Huh-7 and Hep3B cells (Figure 3F). These data suggested that paclitaxel exerted its function by regulating circ-BIRC6 in HCC cells in vitro.
Knockdown of Circ-BIRC6 and Paclitaxel Treatment Inhibited Tumor Growth in vivo
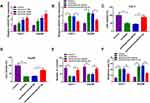
To explore whether circ-BIRC6 and paclitaxel have the same effect in vivo, we established a xenograft model. We observed that circ-BIRC6 knockdown or paclitaxel treatment inhibited tumor volume and weight in the xenograft model and combination of circ-BIRC6 interference and paclitaxel treatment further suppressed tumor growth (Figure 4A and B). Moreover, the expression of circ-BIRC6 was decreased in sh-circ-BIRC6 or paclitaxel group, and its expression was further reduced in sh-circ-BIRC6 + paclitaxel group (Figure 4C). Consistent with in vitro assays, these results demonstrated that circ-BIRC6 downregulation and paclitaxel treatment could inhibit tumor growth in vivo.
Circ-BIRC6 Directly Interacted with miR-877-5p in HCC Cells
Given the alteration of circ-BIRC6 and miR-877-5p in HCC tissues and cell lines, as well as the negative correction between circ-BIRC6 and miR-877-5p expression, we wondered whether the effect of circ-BIRC6 was mediated by miR-877-5p through complementary binding sites. Through web-based tool starBase v2.0, miR-877-5p was predicted to have complementary bases pairing with circ-BIRC6 (Figure 5A). To verify the direct binding between circ-BIRC6 and miR-877-5p, luciferase reporter plasmids containing the WT or MUT in circ-BIRC6 were constructed and co-transfected with miR-877-5p or miR-NC into Huh-7 and Hep3B cells. The data showed that miR-877-5p led to an apparent decrease in luciferase activity of WT-circ-BIRC6 relative to the miR-NC group, but not that of MUT-circ-BIRC6 (Figure 5B and C). qRT-PCR results indicated that si-circ-BIRC6 reduced the expression of circ-BIRC6, and transfection of circ-BIRC6 increased the expression of circ-BIRC6, suggesting that si-circ-BIRC6 and circ-BIRC6 were successfully transfected into Huh-7 and Hep3B cells (Figure 5D). The regulatory effect of circ-BIRC6 on miR-877-5p expression was investigated by qRT-PCR. The data indicated that knockdown of circ-BIRC6 promoted the expression of miR-877-5p, and overexpression of circ-BIRC6 presented an opposite effect (Figure 5E). In addition, interference of miR-877-5p attenuated si-circ-BIRC6-mediated promotion of miR-877-5p expression (Figure 5F). Next, we explored whether the circ-BIRC6-mediated function was regulated by miR-877-5p. As shown in Figure 5G and H, knockdown of circ-BIRC6 repressed cell viability and colony formation, while the silence of miR-877-5p abated these effects. Furthermore, the downregulation of circ-BIRC6 accelerated cell apoptosis, which was reversed by the interference of miR-877-5p (Figure 5I). Overall, these results elucidated that knockdown of circ-BIRC6 inhibited HCC tumorigenesis by up-regulating miR-877-5p.
Paclitaxel Repressed HCC Tumorigenesis via Up-Regulating miR-877-5p
Next, we explored the connection between miR-877-5p and paclitaxel in HCC cells. The results from qRT-PCR showed that paclitaxel exposure dose-dependently increased the expression of miR-877-5p (Figure 6A). Moreover, we found that the downregulation of miR-877-5p abated paclitaxel’ promotion effect on expression of miR-877-5p (Figure 6B). Additionally, silencing miR-877-5p reversed the suppressive effect of paclitaxel on cell viability and colony formation (Figure 6C-E). Also, interference of miR-877-5p abolished the pro-apoptosis effect induced by paclitaxel in Huh-7 and Hep3B cells (Figure 6F). Therefore, we concluded that paclitaxel exerted its effect via modulating miR-877-5p in HCC cells.
YWHAZ Was a Direct Target of miR-877-5p in HCC Cells
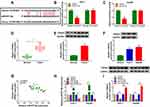
Considering that miRNAs exert their biological roles through binding to the 3ʹUTR of specific target genes, we further predicted the candidate targets of miR-877-5p by starBase v2.0 tool. As displayed in Figure 7A, YWHAZ contained miR-877-5p binding sites in the 3ʹUTR. We performed a dual-luciferase reporter assay to validate this prediction. The results revealed that luciferase activity of YWHAZ 3ʹUTR-WT was markedly reduced in Huh-7 and Hep3B cells after transfection with miR-877-5p compared with the miR-NC group, while no difference was found in the mutant group (Figure 7B and C). Next, we analyzed the expression of YWHAZ in HCC tissues and cells. The results indicated that YWHAZ mRNA level and protein level were increased in HCC tissues compared to normal tissues (Figure 7D and E). Likewise, the protein level of YWHAZ was also elevated in HCC cells compared with THLE-2 cells (Figure 7F). Moreover, we found that the YWHAZ mRNA level was inversely correlated with miR-877-5p expression in HCC tissues (Figure 7G) (r=−0.6372, p<0.001). QRT-PCR analysis showed that miR-877-5p was over-expressed in Huh-7 and Hep3B cells transfected with miR-877-5p compared to control or miR-NC group, and its expression was reduced in those cells transfected with anti-miR-877-5p, suggesting that transfection of miR-877-5p and anti-miR-877-5p was successful (Figure 7H). Besides, we observed that overexpression of miR-877-5p reduced the protein abundance of YWHAZ, and knockdown of miR-877-5p increased the protein level of YWHAZ (Figure 7I). In a word, these data indicated that YWHAZ directly interacted with miR-877-5p.
Circ-BIRC6 Acted as a Molecular Sponge of miR-877-5p to Regulate YWHAZ in HCC Cells
To explore the effects of circ-BIRC6 and miR-877-5p on the expression of YWHAZ, Huh-7 and Hep3B cells were transfected with si-NC, si-circ-BIRC6, si-circ-BIRC6 + anti-miR-NC, or si-circ-BIRC6 + anti-miR-877-5p. Western blot assay presented that interference of circ-BIRC6 reduced the protein expression of YWHAZ, which was abolished by downregulating miR-877-5p (Figure 8A). Next, we analyzed whether paclitaxel could regulate YWHAZ expression. As displayed in Figure 8B, paclitaxel treatment inhibited the protein expression of YWHAZ, while this effect was reversed by the addition of circ-BIRC6. Thus, our data proved that paclitaxel exerted its effect by modulating circ-BIRC6/miR-877-5p/YWHAZ axis.
Discussion
HCC is common and aggressive, and it is related to a large number of cancer-related deaths in the world.20 Emerging evidence shows that circRNAs play functional roles in the pathogenesis of tumors and also offer novel insights into the biology of cancer progression.21,22 Changes in circRNA expression have been found in various cancer cells exposed to radiotherapy or chemotherapy.23,24 Many reports have shown that paclitaxel-based chemotherapy is especially effective in multiple cancer cases.5,25 Previous studies have demonstrated that paclitaxel sensitivity may be linked to the expression of circCELSR1 in ovarian cancer,26 hsa_circ_0002483 in lung cancer,27 and circ-PVT1 in gastric cancer.28 However, there is no study on the association between circRNAs and paclitaxel in HCC.
Some studies have proven that paclitaxel could inhibit the progression of HCC through inhibiting growth and inducing cell cycle arrest and apoptosis.29,30 Here, we found that paclitaxel treatment suppressed cell growth and accelerated apoptosis in HCC cells. A previous study showed that circ-BIRC6 was highly expressed in HCC tissues and cells, and circ-BIRC6 knockdown inhibited the progression of HCC via regulating the miR-3918/Bcl2 axis.13 Moreover, miR-877-5p was reported to be downregulated and play an anti-cancer role in many cancers, such as cervical cancer,31 glioblastoma,32 renal cell carcinoma,33 and colorectal cancer,34 as well as HCC.17 In agreement with these findings, we also observed that circ-BIRC6 expression was enhanced, and the miR-877-5p level was reduced in HCC tissue samples and cells. Next, we explored whether the effect of paclitaxel was regulated by circ-BIRC6 and miR-877-5p. We found that paclitaxel treatment dose-dependently decreased circ-BIRC6 expression in HCC cells, while increased the abundance of miR-877-5p in HCC cells. Moreover, overexpression of circ-BIRC6 or silence of miR-877-5p decreased the suppressive effect of paclitaxel on HCC tumorigenesis. Consistent with our results, a previous study proved that miR-877 level was elevated in paclitaxel-treated HCC cells, and its restoration partially restored paclitaxel sensitivity.35 Moreover, we uncovered that paclitaxel treatment and knockdown of circ-BIRC6 inhibited tumor growth in vivo. These data suggested that paclitaxel inhibited the progression of HCC through down-regulating circ-BIRC6 and up-regulating miR-877-5p.
Accumulating evidence shows that some circRNAs participate in tumorigenesis through functioning as sponges for miRNAs to modulate gene transcription or protein translation.36 Interestingly, miR-877-5p was identified as a direct target of circ-BIRC6, and interference of miR-877-5p reversed the inhibitory effect of circ-BIRC6 knockdown on HCC tumorigenesis, implying that circ-BIRC6 exerted its function by sponging miR-877-5p. To further elucidate the regulatory mechanism of miR-877-5p in HCC, we searched the possible downstream targets of miR-877-5p using the starBase v2.0. We uncovered that YWHAZ was a downstream target of miR-877-5p. YWHAZ has been demonstrated to participate in diverse physiological and pathological processes as well as drug resistance in human cancer cells.37,38 More importantly, some reports indicated that YWHAZ was overexpressed and acted as an oncogene in HCC.39–41 In line with these results, we observed that YWHAZ was increased in HCC tissue samples and cells. Additionally, circ-BIRC6 positively regulated YWHAZ by sponging miR-877-5p, and paclitaxel inhibited the expression of YWHAZ by regulating circ-BIRC6 expression. These findings collectively indicated that paclitaxel repressed the progression of HCC through regulating circ-BIRC6/miR-877-5p/YWHAZ axis.
In summary, we demonstrated that paclitaxel inhibited the expression of circ-BIRC6 and YWHAZ while promoted expression of miR-877-5p. Paclitaxel suppressed HCC tumorigenesis by modulating circ-BIRC6/miR-877-5p/YWHAZ axis (Figure 8C). These findings may serve as guidelines for the development of paclitaxel-based chemotherapy for the treatment of HCC.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Suizhou hospital, Hubei University of Medicine. Written informed consent was obtained from all enrolled patients.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
No funding was received.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Jinjuvadia R, Salami A, Lenhart A, Jinjuvadia K, Liangpunsakul S, Salgia R. Hepatocellular carcinoma: a decade of hospitalizations and financial burden in the United States. Am J Med Sci. 2017;354(4):362–369. doi:10.1016/j.amjms.2017.05.016
3. Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485. doi:10.1200/JCO.2008.20.7753
4. Jin C, Li H, He Y, et al. Combination chemotherapy of doxorubicin and paclitaxel for hepatocellular carcinoma in vitro and in vivo. J Cancer Res Clin Oncol. 2010;136(2):267–274. doi:10.1007/s00432-009-0658-5
5. Markman M, Mekhail TM. Paclitaxel in cancer therapy. Expert Opin Pharmacother. 2002;3(6):755–766. doi:10.1517/14656566.3.6.755
6. Wan Y-F, Guo X-Q, Wang Z-H, Ying K, Yao M-H. Effects of paclitaxel on proliferation and apoptosis in human acute myeloid leukemia HL-60 cells. Acta Pharmacol Sin. 2004;25(3):378–384.
7. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
8. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi:10.1261/rna.047126.114
9. Zhang J, Zhao X, Zhang J, Zheng X, Li F. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem Biophys Res Commun. 2018;501(2):428–433. doi:10.1016/j.bbrc.2018.05.006
10. Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19. doi:10.1186/s12943-018-0771-7
11. Liang H-F, Zhang X-Z, Liu B-G, Jia G-T, Li W-L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566.
12. Liu L, Liu F-B, Huang M, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18(6):580–586. doi:10.1016/j.hbpd.2019.03.003
13. Yang G, Wang X, Liu B, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle. 2019;18(9):976–989. doi:10.1080/15384101.2019.1601477
14. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
15. Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161.
16. Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610.
17. Yan T, Qiu C, Sun J, Li W. MiR-877-5p suppresses cell growth, migration and invasion by targeting cyclin dependent kinase 14 and predicts prognosis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(10):3038–3046. doi:10.26355/eurrev_201805_15061
18. Fan Z, Cui H, Yu H, et al. MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis. 2016;5(2):e197e197. doi:10.1038/oncsis.2016.1
19. Zhao J-F, Zhao Q, Hu H, et al. The ASH1-miR-375-YWHAZ signaling axis regulates tumor properties in hepatocellular carcinoma. Molecular Therapy-Nucleic Acids. 2018;11:
20. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359E386. doi:10.1002/ijc.29210
21. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):1–10. doi:10.1186/s12943-018-0934-6
22. Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2. doi:10.1186/s13045-016-0370-2
23. Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9(9):1175–1188. doi:10.2217/epi-2017-0055
24. Chen Y, Yuan B, Wu Z, Dong Y, Zhang L, Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. 2017;629:35–42. doi:10.1016/j.gene.2017.07.078
25. Reichman BS, Seidman AD, Crown J, et al. Paclitaxel and recombinant human granulocyte colony-stimulating factor as initial chemotherapy for metastatic breast cancer. J Clin Oncol. 1993;11(10):1943–1951. doi:10.1200/JCO.1993.11.10.1943
26. Zhang S, Cheng J, Quan C, et al. circCELSR1 (hsa_circ_0063809) contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Molecular Therapy-Nucleic Acids. 2020;19:
27. Li X, Yang B, Ren H, et al. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10(12):1–12. doi:10.1038/s41419-019-2180-2
28. Liu Y-Y, Zhang L-Y, Du W-Z. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR-124-3p. Biosci Rep. 2019;39:12. doi:10.1042/BSR20193045
29. Chun E, Lee K-Y. Bcl-2 and Bcl-xL are important for the induction of paclitaxel resistance in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2004;315(3):771–779. doi:10.1016/j.bbrc.2004.01.118
30. Okano J-I, Nagahara T, Matsumoto K, Murawaki Y. The growth inhibition of liver cancer cells by paclitaxel and the involvement of extracellular signal-regulated kinase and apoptosis. Oncol Rep. 2007;17(5):1195–1200.
31. Liang J, Zhang S, Wang W, et al. Long non-coding RNA DSCAM-AS1 contributes to the tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3 axis. Biosci Rep. 2020;40(1):BSR20192061. doi:10.1042/BSR20192061
32. Xie H, Shi S, Chen Q, Chen Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathology-Research and Practice. 2019;215(8):152476. doi:10.1016/j.prp.2019.152476
33. Shi Q, Xu X, Liu Q, Luo F, Shi J, He X. MicroRNA‑877 acts as a tumor suppressor by directly targeting eEF2K in renal cell carcinoma. Oncol Lett. 2016;11(2):1474–1480. doi:10.3892/ol.2015.4072
34. Zhang L, Li C, Cao L, et al. microRNA-877 inhibits malignant progression of colorectal cancer by directly targeting MTDH and regulating the PTEN/Akt pathway. Cancer Manag Res. 2019;11:2769. doi:10.2147/CMAR.S194073
35. Huang X, Qin J, Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int J Clin Exp Pathol. 2015;8(2):1515.
36. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
37. Li Y, Zou L, Li Q, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16(2):214. doi:10.1038/nm.2090
38. Nishimura Y, Komatsu S, Ichikawa D, et al. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer. 2013;108(6):1324–1331. doi:10.1038/bjc.2013.65
39. Jiang X, Wu J, Zhang Y, et al. MiR-613 functions as tumor suppressor in hepatocellular carcinoma by targeting YWHAZ. Gene. 2018;659:168–174. doi:10.1016/j.gene.2018.03.036
40. Chen M, Hu W, Xiong C-L, et al. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget. 2016;7(49):80751. doi:10.18632/oncotarget.13037
41. Wei G, Hu M, Zhao L, Guo W. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5158–5167. doi:10.26355/eurrev_201906_18180
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.