Back to Journals » OncoTargets and Therapy » Volume 14
P4HA1 Down-Regulation Inhibits Glioma Invasiveness by Promoting M1 Microglia Polarization
Authors Wang Q , Zhang J , Fang S, Wang J, Han X, Liu F , Jin G
Received 31 December 2020
Accepted for publication 16 February 2021
Published 8 March 2021 Volume 2021:14 Pages 1771—1782
DOI https://doi.org/10.2147/OTT.S299977
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Qiyan Wang,* Junwen Zhang,* Sheng Fang, Jialin Wang, Xiangming Han, Fusheng Liu, Guishan Jin
Brain Tumor Research Center, Beijing Neurosurgical Institute, Beijing Laboratory of Biomedical Materials, Beijing Tiantan Hospital Affiliated to Capital Medical University, Beijing, 10070, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guishan Jin; Fusheng Liu
Brain Tumor Research Center, Beijing Neurosurgical Institute, Beijing Laboratory of Biomedical Materials, Beijing Tiantan Hospital Affiliated to Capital Medical University, No. 119 Nansihuanxilu, Fengtai District, Beijing, 100070, People’s Republic of China
Tel +86-10-59975626
Email [email protected]; [email protected]
Background: Polarization of microglia cells in the glioma microenvironment is closely related to the malignant progression and invasion of gliomas. Prolyl 4-hydroxylase subunit α 1 (P4HA1) is the rate-limiting subunit of prolyl 4-hydroxylase (P4H). In previous studies, we showed that P4HA1 could promote the proliferation, migration, and invasion of glioma cells, but the specific mechanisms through which this occurs have not been fully elucidated.
Materials and Methods: Interactions between glioma and microglia cells were analyzed using bioinformatics. Then, co-culture models were used to obtain conditioned media. To characterize microglial cell polarization, we used PCR and immunofluorescence. Proliferation and invasion assays were used to explore the biological behavior of glioma cells affected by microglia. Finally, marker expression was detected using immunohistochemistry in glioblastoma multiform (GBM) specimens.
Results: Knockdown of P4HA1 resulted in reduced chemotaxis of microglia toward GBM cells and increased polarization of microglia toward the M1 phenotype. The changed microglial polarization state, in turn, inhibited the proliferation and invasion of GBM cells. Moreover, in GBM tissue specimens, the P4HA1 expression level is negatively correlated with that of the CD86 microglia M1-specific marker.
Conclusion: Our results show that P4HA1 promotes immunosuppressive microenvironment formation by cross-talk between GBM and microglia cells and indirectly increases the aggressiveness of GBM.
Keywords: glioma, microglia, P4HA1, polarization, invasiveness
Introduction
Glioblastoma multiform (GBM) is the most common primary brain tumor in adults, accounting for about 1% of new cancer cases worldwide annually.1 The median overall survival time of patients with GBM is less than 15 months, even with the most aggressive treatments being developed and used in the past decades.2,3 To date, there is no effective treatment regimen for patients with GBM. In recent years, encouraging progress has been made in immunotherapy-based treatments of other tumors. This, combined with a deeper understanding of the complicated interaction between glioma cells and the immune system, means that immunotherapy strategies targeting tumor-associated macrophages (TAMs) in the glioma microenvironment have become a research hotspot.4
TAMs are the major infiltrating immune cell population in gliomas. TAMs are composed of the innate macrophages of the central nervous system (CNS), namely microglia cells and bone marrow-derived macrophages. As the only resident macrophage in the brain, microglia exert immune surveillance functions and maintain the homeostasis of the CNS.5,6 In physiological conditions, microglia typically assume an M0-type expression profile. However, microglia/macrophages are polarized into activation phenotypes under pathological conditions. In such conditions, microglia/macrophages are roughly divided into two phenotypes, the classically activated pro-inflammatory M1 phenotype and the alternatively activated anti-inflammatory M2 phenotype.7,8 When inflammation occurs, microglia cells are activated by interferon (IFN) or lipopolysaccharide (LPS). This produces M1 polarization of microglia with high expression of CD16, CD32, TNF-α, MCP-1, CD86, CD64, CD80, and other surface markers, generally presenting anti-inflammatory and anti-tumor activities.8,9 During tumorigenesis, glioma cells secret soluble factors including MCP-1 (CCL2), SDF-1 (CXCL12), M-CSF (CSF-1), GM-CSF, and EGF, and recruit microglia cells and transform them to the M2 phenotype. M2-type microglia can secret TGF-β, ST1, EGF, IL-6, and IL-1β to promote glioma growth and invasion and express some surface markers at high levels, including CD163, CD206, CD45, CD11b, and CD23.9–14 Furthermore, M2 type microglia dominate the glioma microenvironment, producing soluble factors to induce immunosuppression in addition to cell-to-cell contact communication, and promoting glioma malignant proliferation and metastasis. In contrast, M1-type microglia appear to do the opposite in the inflammatory state.9,14 TAMs are considered the key driver of glioma local immunosuppression, tumor progression, and resistance to immunomodulating treatment strategies.11,15
Prolyl 4-hydroxylase subunit α1 (P4HA1) is the rate-limiting subunit of prolyl 4-hydroxylase (P4H). P4H is an essential enzyme for procollagen hydroxylation and collagen synthesis and secretion.16 P4HA1 can promote the malignant progression of breast, prostate, and pancreatic cancers, but the mechanism remains unclear.17–19 Previously, we found that P4HA1 promotes the invasive growth of gliomas.20 We used The Cancer Genome Atlas (TCGA) dataset to investigate P4HA1 gene expression profiles and found that P4HA1 expression significantly positively correlated with the pathological grade of gliomas and was distinctly associated with TAM polarization. To explore whether P4HA1 promotes glioma invasiveness by changing the microglia polarization state in the tumor microenvironment, we co-cultured glioma cells and microglial cells in vitro and then measured the expression of P4HA1, M1-specific, and M2-specific phenotypic markers in surgically resected glioma specimens in vivo, attempting to figure out the role of P4HA1 in the cross-talk between GBM cells and the polarization of microglial cells.
Materials and Methods
Materials
U251 and U87 glioma cell lines were obtained from the Cell Resource Center, Peking Union Medical College (National Infrastructure of Cell Line Resources, NSTI). These cell lines were authenticated by short tandem repeat (STR) profiles. The human HMC-3 microglia cell line was purchased from the American Type Culture Collection (ATCC, USA). P4HA1 knockdown (sh-P4HA1) and empty vector (sh-Ctrl) U251 and U87 cells were all established in our laboratory. TRIzol total RNA isolation reagent was purchased from Invitrogen, USA. cDNA reverse transcription kit was purchased from Promega, USA. PowerUp™ SYBR™ Green Master Mix was purchased from Applied Biosystems (ABI, USA). Rabbit Anti-Human P4HA1 antibody (12,658-1-AP), Rabbit Anti-Human CD86 antibody (13,395-1-AP), and Mouse Anti-Human CD206 antibody (60,143-1-Ig) were purchased from ProteinTech, China. Secondary antibodies for immunofluorescence analyses, Anti-rabbit IgG (H+L) (Alexa Fluor® 488 Conjugate) and Anti-mouse IgG (H+L) (Alexa Fluor® 594 Conjugate), were purchased from Cell Signaling Technology (CST, USA). Goat anti-rabbit and goat anti-mouse horseradish peroxidase (HRP) were purchased from Neobioscience, China. Cell Counting Kit-8 (CCK-8) was purchased from Dojindo, Japan. Real-time quantitative PCR analysis was performed using a QuantStudio™ 5 Real-time PCR System (ABI, USA).
Cell Culture and Preparation of Conditioned Medium
U251, U87, and HMC-3 cells were cultured in Dulbecco’s Modified Eagle medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Ever Green, China) and 50 U/mL penicillin-streptomycin solution (Gibco, USA) at 37°C in a 5% CO2 humidified cell incubator (Thermo, Germany).
The co-culture conditioned medium was collected by co-culturing GBM and microglia cells in 6-well Transwell plates (pore diameter 0.4 μm). Conditioned medium was also obtained from glioma cells alone, without co-culture with microglia cells. In detail, when co-cultured with microglia cells, U251 (Control), U251 with knocked down P4HA1 (sh-P4HA1), and U251 with empty vector (sh-Ctrl) were each seeded in the upper Transwell chambers at a density of 2×104/mL/well. The lower chambers were seeded with HMC-3 cells at a density of 105/2mL/well. Resting HMC-3 cells were used as the blank group, with no glioma cells in the upper chambers. To obtain the conditioned medium for glioma cells alone, the number of glioma cells seeded in each group and the culture time remained the same, but the lower chambers of all groups did not contain HMC-3 cells. After 72 hours of culture, the medium was collected from each group and filtered using a 0.2 μm filter. All U87 groups were treated in the same way.
Immunohistochemistry and Immunofluorescence
After fixing with 10% formaldehyde for 48 hours, glioma specimens were embedded in paraffin. Tumor tissues were sectioned at a thickness of 4 μm, and deparaffinization, antigen retrieval, and nonspecific-binding blocking were performed using standard procedures. Rabbit anti-human P4HA1 (1:200), rabbit anti-human CD86 (1:200), and mouse anti-human CD206 (1:400) primary antibodies were added and incubated overnight at 4°C. 3,3ʹ-Diaminobenzidine (DAB) staining and hematoxylin counterstaining were performed the following day, and the slides were dehydrated and sealed with neutral resin. Sections were examined at 200× magnification, and representative images were obtained from four randomly selected fields of a section adjacent to that identified by hematoxylin and eosin (HE). Staining results were quantified using Image J.
Immunofluorescence staining of microglia polarization markers was grouped as above. In the Transwell co-culture systems, 14-mm circular slides were placed in the lower chambers before seeding microglia cells. After co-culturing, the slides were taken out, washed with prewarmed phosphate-buffered saline (PBS), and fixed in 4% paraformaldehyde for 20 minutes. The slides were then blocked with 5% bovine serum albumin for 1 hour at room temperature. Rabbit anti-human CD86 (1:400) and mouse anti-human CD206 (1:400) primary antibodies were added and incubated at 4°C overnight. Slides were washed three times with PBS and then incubated with fluorescent secondary antibody diluted at 1:2000 for 1 hour at room temperature. Finally, after three washes with PBS plus Tween 20, slides were sealed using antifade mountant with 4ʹ,6-diamidino-2-phenylindole (DAPI). Four randomly selected fields were photographed under a fluorescence microscope, and the mean fluorescence intensity was analyzed using Image J.
Cell Proliferation Assays
U251 and U87 cells growing at exponential phase were digested and counted, then suspended with the conditioned medium in 96-well plates at a density of 5×103/100μL/well and cultured at 37°C with 5% CO2. After 48 hours, the supernatants were discarded, and 100 μL of CCK-8 reagent, diluted at 1:10 in DMEM, was added. The reaction was incubated for 120 minutes and the Optical Density (OD) at 450 nm was determined using a microplate reader. Four duplicates were completed for each group, and each test was repeated at least three times.
Cell Chemotaxis Assays
Exponentially growing cells were digested, counted, and seeded. In brief, HMC-3 cells were resuspended with serum-free DMEM and seeded into the upper chambers of the Transwell system at a density of 2×104/200μL/well, while U251 (Control), U251 with P4HA1 knocked down (sh-P4HA1), and U251 with empty vector (sh-Ctrl) were resuspended in DMEM supplemented with 10% FBS and seeded in the lower chambers at a density of 6×104/600μL/well. A cell-free group (Blank) was used to exclude the influence of serum. After incubation in the cell incubator for 16 hours, the cells on the upper surface of the membrane were wiped with cotton swabs. The migrated cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature and then stained with 3% (W/V) crystal violet for 20 minutes. Each group was set with three duplicates. Four random fields of adherent cells in each well were photographed and counted using ImageJ software. All U87 groups were treated the same way.
Cell Invasion Assays
Transwell assays were used to determine the influence of microglia polarization on glioma cell invasion ability. The upper Transwell chambers were coated with Matrigel, with a dilution ratio of 1:6 in DMEM. U251 and U87 cells were cultured in the co-culture conditioned medium for 48 hours. After digesting and counting, cells were resuspended in serum-free DMEM and seeded into the upper chambers at a density of 5×104/200μL/well. DMEM containing 10% FBS was added to the lower chambers. Each group was set with three duplicates. After incubation in the cell incubator for 24 hours, the cells were dyed and counted in ways as chemotaxis assays were performed.
Total RNA Isolation and qRT-PCR Analyses
Total RNA was isolated using the TRIzol reagent and reversely transcribed to produce cDNA. Reverse-transcription PCR (RT-PCR) was performed in standard mode, the GAPDH housekeeping gene was used as the internal control, and the -ΔΔCt algorithm was used for relative quantification. Each group was assigned to four duplicates, and each test was repeated at least three times. The primers used were synthesized by Beijing Tianyihuiyuan Biotechnology Co. Ltd, and their sequences are described in Table 1.
 |
Table 1 qPCR Primers Used in the Experiment |
Bioinformatics Analysis
The gene expression profiles of glioma of different pathological grades were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). The GBM array was divided into High and Low expression groups based on P4HA1 expression levels, and CIBERSORTx (https://cibersortx.stanford.edu/) was used to analyze the relative abundance of the tumor-infiltrating immune cell population.21
Statistical Analyses
GraphPad Prism 8 software was used for statistical analyses. Differences between groups were analyzed using t-test, one-way ANOVA, or two-way ANOVA. P-value ≤ 0.05 was considered statistically significant.
Results
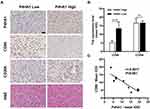
P4HA1 is Highly Expressed in HGG and Negatively Correlated with M1 Polarization of TAMs
P4HA1 mRNA expression levels were evaluated in different WHO grade gliomas using gene expression profile data from the TCGA database. P4HA1 was significantly up-regulated in high-grade gliomas (HGG) (WHO III and WHO IV) compared to low- grade gliomas (HGG) (WHO I and WHO II) (P < 0.0001, Figure 1A). To determine the relationship between P4HA1 expression and TAM polarization in HGG, especially in GBM, CIBERSORTx methodology was used to determine the relative abundance of M1- or M2-polarized TAMs in GBM. RNA-seq matrix profiles, distributed in quartiles to the high and the low P4HA1 expression groups (Figure 1B), were uploaded to CIBERSORTx to relatively quantify the abundance of immune cell infiltration (Figure 1C and D).21 The infiltration of M1 TAMs in the P4HA1 low-expression group was significantly higher than that in the P4HA1 high-expression group. This suggests that P4HA1 expression levels negatively correlate with M1 polarization of TAMs in GBM (P < 0.001). However, P4HA1 expression did not statistically correlate with M2 polarization of TAMs (P > 0.05).
P4HA1 Knockdown Weakens the Chemotaxis of Microglia Toward GBM Cells
We next sought to investigate whether the P4HA1 expression affects microglial chemotaxis toward GBM cells using the Transwell migration assay. In brief, Transwell chambers were divided into four groups. All four groups were inoculated with HMC-3 microglia cells in the upper chambers, while the lower chambers were seeded with GBM cells. The blank group, containing no cells, was also set at the same time. After 17 hours of culture, HMC-3 cells that had passed through the membranes were stained and counted (Figure 2A). There was no statistically significant difference between the empty vector group and the control group in either U251 or U87 cells (P < 0.05). Comparison of these two groups with the blank control group revealed an obvious increase in HMC-3 cell migration (P < 0.0001 for both) (Figure 2B). P4HA1 knockdown distinctly decreased HMC-3 cell migration (vs Control and sh-Ctrl, both P < 0.0001) and tended to restore the migration number to a low level similar to that of the blank group (P > 0.05). These results indicate that GBM cells have chemotaxis on microglia and that the vector itself does not disturb this chemotaxis. Notably, P4HA1 down-regulation can impede microglial chemotaxis toward GBM cells and restore it to the unaffected levels (vs blank, P > 0.05).
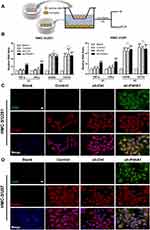
Knockdown of P4HA1 in GBM Facilitates M1 Polarization of Microglia
To verify whether GBM P4HA1 expression levels affect the polarization state of microglia cells, we co-cultured U251 and U87 GBM cells with HMC-3 microglia cells in the Transwell 6-well plate. After 72 hours, transcriptome analysis and immunofluorescence staining were used to detect microglia polarization markers (Figure 3A). The results showed that mRNA levels of TNF-α and IFN-γ M1 phenotype markers in the Control and sh-Ctrl groups did not significantly differ from those in the blank group (P > 0.05), and were significantly increased in the sh-P4HA1 group (P < 0.0001).
The mRNA levels of the CD206 and CD163 M2-type surface markers did not differ between the Control, sh-Ctrl, and sh-P4HA1 groups, but were significantly increased in three groups compared with the blank group (P < 0.0001, Figure 3B). These results are consistent with those observed by immunofluorescence staining (Figure 3C and D), which showed that the CD86 M1 surface marker was only observed in the sh-P4HA1 group, while the CD206 M2 surface marker was expressed in all groups except the blank group. These results suggest that GBM cells are inclined to maintain microglia cells in an M2-specific phenotype. Although P4HA1 knockdown did not change the M2-related expression profile of microglia, it induced M1-related marker up-regulation, including CD86, TNF-a, and IFN-γ. Together, these data indicate that P4HA1 participates in signaling pathways that function to regulate the M1-related phenotype of microglia.
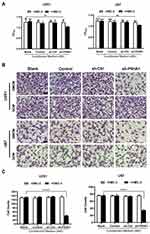
M1 Polarization of Microglia Inhibits GBM Proliferation and Invasiveness
To determine if the polarization state of microglia affects GBM cell proliferation, we collected the conditioned medium of each group and respectively applied them to U251 and U87 cells during exponential growth. We then used CCK-8 assays to assess U251 and U87 cell proliferation. Conditioned medium derived from the sh-P4HA1 group, in which GBM cells with knocked-down P4HA1 were co-cultured with HMC-3 cells, significantly inhibited the proliferative activity of U251 and U87 GBM cells compared with control groups, including the GBM cells conditioned medium (GCM) and the co-culture conditioned medium with HMC-3 (GMCM). This inhibitory effect was more obvious in U87 GBM cells (Figure 4A).
Transwell assays were used to explore the influence of the conditioned medium on GBM cell invasiveness (Figure 4B). The sh-P4HA1 group conditioned medium, in which GBM cells were co-cultured with HMC-3 microglia cells, exhibited a more substantial inhibitory effect on GBM cell invasive ability than did the other groups, including GCM and GMCM. The inhibitory effect was consistent in U251 and U87 GBM cell lines (P < 0.0001) (Figure 4C).
Given the above data, we propose that GBM cells with low levels of P4HA1 induce microglia polarization toward the M1 phenotype through interaction with microglia cells, thus playing an inhibitory role in tumor growth and invasion.
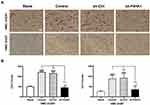
P4HA1 Expression Negatively Correlates with CD86 M1 Marker Expression in GBM Tissue Specimens
Immunohistochemical staining was used to detect P4HA1, CD86, and CD206 expression levels in ten human GBM tissue specimens (Figure 5A). Image-Pro Plus 6.0 software was used to divide staining results into low and high groups based on the mean IOD values of P4HA1 protein. The P4HA1 low expression group exhibited distinctly higher levels of CD86 (P < 0.0001), but no difference was observed in CD206 (P > 0.05) (Figure 5B). Additional correlation analysis confirmed that the P4HA1 expression was negatively correlated with the CD86 in microglia cells (P < 0.001) (Figure 5C).
Discussion
Standard treatments for GBM patients include surgical resection, radiation therapy (RT), and temozolomide chemotherapy. The low efficacy of therapy can be attributed to the highly aggressive growth pattern of GBM. Even with total radical excision, the tumors are not always completely eradicated. Moreover, GBM retains a high inherent level of heterogeneity between different GBM and within individual GBM masses. As a result, the signaling pathways involved in drug resistance and chemoradiotherapy resistance in GBM are extremely complex. Additionally, GBM harbors a complicated tumor microenvironment (TME). Recent experimental and clinical studies have shown that the TME plays a vital role in promoting tumorigenesis and drug resistance in GBM. Targeting the immunosuppressive TME provides new ideas and approaches for GBM treatment.9,11
In GBM, the TME mainly includes non-neoplastic cells, primarily infiltrating inflammatory cells and intrinsic cells in brain tissues including neurons, astrocytes, microglia, and vascular endothelial cells; extracellular matrix components, including the stromal structure around tumor cells, which are rich in hyaluronic acid; and soluble signaling factors, such as cytokines, growth factors, and specific glioma proteins.22–24 When glioma occurs, a complicated network of intercellular signalings is established through a series of soluble molecules, including cytokines, growth factors, and certain glioma proteins. Together with glioma cells, glioma stem-like cells, and non-neoplastic cell components such as astrocytes, microglia, and infiltrating immune cells, an immunosuppressive microenvironment is created to promote the malignant progression of gliomas.25–27 The density of infiltrating TAMs is positively associated with the malignant characteristics, pathologic grades, and invasive properties of GBM. In GBM, the proportion of TAMs can be as high as 30–50% of the cell components in the tumor mass.23,24,28 In recent years, TAMs have become a popular therapeutic target, and further studies on the mechanisms of the interplay between tumor cells and non-neoplastic cells are expected to reveal new therapeutic strategies. Using the TCGA database to analyze the association between P4HA1 and other proteins in glioma, we found that P4HA1 expression was highly positively correlated with pathological grade. This is consistent with results previously reported by our team and further confirms the positive correlation between P4HA1 and the malignant progression and invasive abilities of gliomas.20 Meanwhile, we also found a correlation between P4HA1 expression levels and the polarization state of TAMs. Indeed, the P4HA1 expression level was negatively correlated with the M1 phenotype, and no correlation was observed between P4HA1 expression and M2-type polarization (Figure 1C and D), suggesting that P4HA1 may be involved in the interplay between glioma cells and TAMs in the glioma microenvironment.
To evaluate whether P4HA1 levels in GBM have an impact on the recruitment and chemotaxis of microglia cells, we conducted Transwell assays. We found that down-regulation of P4HA1 in U251 and U87 GBM cell lines can significantly reduce chemotaxis of the HMC-3 microglia cell line (Figure 2). This indicates that P4HA1 is involved in the recruitment of microglial cells and accelerates chemotaxis, thus constructing a microenvironment conducive to the malignant behaviors of glioma. Since TAMs infiltration is considered one of the driving factors for the development of glioma malignancy, it may also represent a promising therapeutic strategy to hinder TAMs’ infiltration or accelerate their death.15
TAMs are highly plastic and maintain the ability to switch between the anti-tumor M1 and the pro-tumor M2 phenotypes.29,30 To research whether the polarization state of recruited microglia is affected by P4HA1 expression levels in GBM, we co-cultured glioma cells and microglia cells. We found that knockdown of P4HA1 significantly up-regulated the expression level of M1 markers in microglia, including TNF-α, IFN-γ, and CD86. Still, CD206 and CD163 M2 markers were not affected (Figure 3). Further, in the GBM surgical resection specimens, we confirmed that P4HA1 expression was negatively correlated with the CD86 M1 marker and had no significant correlation with the CD206 M2 marker. We deduced that P4HA1 might change the polarization state of microglial cells to create a favorable microenvironment for malignant glioma progression. At present, several studies have attempted to treat gliomas by polarizing TAMs and maintaining them in the anti-tumor M1 phenotype.30–34 Wang et al used nano-polymer biomaterials to carry IL-12 into the TME to promote TAMs polarization from the M2 to the M1 phenotype.30 Using oncolytic virus expressing IL-12 in conjunction with two immune checkpoint inhibitors, CTLA-4 and PD-1 antibody, Saha et al observed that tumor growth was inhibited in GBM-bearing mice and that the survival time was significantly prolonged.34
To further investigate whether the microglia polarization changes induced by P4HA1 were able to influence GBM cells and change their biological behaviors, cell proliferation and invasion assays were used. We found that after M1 polarization, the secreted proteins of microglia can inhibit proliferative activity and invasion ability of U251 and U87 cells. Our previous studies have shown that P4HA1 knockdown can inhibit the proliferation, migration, and tube formation of glioma or glioma stem-like cells in vitro,20 and inhibit tumorigenesis and prolong survival time in glioma-bearing mice in vivo. These results indicate that levels of P4HA1 in GBM affect the biological behaviors of GBM cells and these effects are deepened through the cross-talk between GBM and microglia cells.
In conclusion, this study preliminarily explored the role of P4HA1 in the cross-talk between GBM cells and microglia in highly malignant GBM. Our results show that P4HA1 has the potential to promote TAM infiltration in GBM and induce the reduction of the anti-tumor M1 phenotype. These two elements help to form the immunosuppressive microenvironment and formidable immune escape. These data have further elucidated the role and function of P4HA1 in the glioma microenvironment, and provide another avenue through which immunotherapy strategies in glioma can be enriched.
Ethical Statement
Human GBM specimens were obtained from the Department of Neurosurgery, Beijing Tiantan Hospital affiliated to Capital Medical University. The histological diagnoses were confirmed as GBM (WHO IV). Tissue collection was approved by the Ethical Committee of Beijing Tiantan Hospital, Capital Medical University. Informed consent for publication was obtained from patients according to the guidelines of the Declaration of Helsinki and its later amendments.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81772671) and the Beijing Laboratory of Biomedical Materials Foundation.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi:10.1056/NEJMoa043330
3. Wen PY, Reardon DA. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12(2):69–70. doi:10.1038/nrneurol.2015.242
4. Poon CC, Sarkar S, Yong VW, Kelly JJP. Glioblastoma-associated microglia and macrophages: targets for therapies to improve prognosis. Brain. 2017;140(6):1548–1560. doi:10.1093/brain/aww355
5. Fu R, Shen Q, Xu P, Luo JJ, Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol Neurobiol. 2014;49(3):1422–1434. doi:10.1007/s12035-013-8620-6
6. Kettenmann H, Kirchhoff F, Verkhratsky A. Microglia: new roles for the synaptic stripper. Neuron. 2013;77(1):10–18. doi:10.1016/j.neuron.2012.12.023
7. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi:10.1016/S1471-4906(02)02302-5
8. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi:10.12703/P6-13
9. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi:10.1038/nn.4185
10. Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216(1):15–24. doi:10.1002/path.2370
11. Prionisti I, Buhler LH, Walker PR, Jolivet RB. Harnessing microglia and macrophages for the treatment of glioblastoma. Front Pharmacol. 2019;10:506. doi:10.3389/fphar.2019.00506
12. Roesch S, Rapp C, Dettling S, Herold-Mende C. When immune cells turn bad—tumor-associated microglia/macrophages in glioma. Int J Mol Sci. 2018;19(2):436. doi:10.3390/ijms19020436
13. Brown NF, Carter TJ, Ottaviani D, Mulholland P. Harnessing the immune system in glioblastoma. Br J Cancer. 2018;119(10):1171–1181. doi:10.1038/s41416-018-0258-8
14. Yin J, Valin KL, Dixon ML, Leavenworth JW; Role of myeloid cells in the immunosuppressive microenvironment in gliomas. The role of microglia and macrophages in CNS homeostasis, autoimmunity, and cancer. J Immunol Res. 2017;2017:1–12. doi:10.1155/2017/5150678
15. Locarno CV, Simonelli M, Carenza C, et al. Role of myeloid cells in the immunosuppressive microenvironment in gliomas. Immunobiology. 2020;225(1):151853. doi:10.1016/j.imbio.2019.10.002
16. Chen L, Shen YH, Wang X, et al. Human prolyl-4-hydroxylase α(i) transcription is mediated by upstream stimulatory factors. J Biol Chem. 2006;281(16):10849–10855. doi:10.1074/jbc.M511237200
17. Xu R. P4HA1 is a new regulator of the HIF-1 pathway in breast cancer. Cell Stress. 2019;3(1):27–28. doi:10.15698/cst2019.01.173
18. Chakravarthi BVSK, Pathi SS, Goswami MT, et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5(16):6654–6669. doi:10.18632/oncotarget.2208
19. Cao XP, Cao Y, Li WJ, Zhang HH, Zhu ZM. P4HA1/HIF1α feedback loop drives the glycolytic and malignant phenotypes of pancreatic cancer. Biochem Biophys Res Commun. 2019;516(3):606–612. doi:10.1016/j.bbrc.2019.06.096
20. Zhou Y, Jin G, Mi R, et al. Knockdown of P4HA1 inhibits neovascularization via targeting glioma stem cell-endothelial cell transdifferentiation and disrupting vascular basement membrane. Oncotarget. 2017;8(22):35877–35889. doi:10.18632/oncotarget.16270
21. Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi:10.1038/s41587-019-0114-2
22. Haddad-Tóvolli R, Dragano NRV, Ramalho AFS, Velloso LA. Development and function of the blood-brain barrier in the context of metabolic control. Front Neurosci. 2017;11:224. doi:10.3389/fnins.2017.00224
23. Kostianovsky AM, Maier LM, Anderson RC, Bruce JN, Anderson DE. Astrocytic regulation of human monocytic/microglial activation. J Immunol. 2008;181(8):5425–5432. doi:10.4049/jimmunol.181.8.5425
24. Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59(8):1169–1180. doi:10.1002/glia.21136
25. Wu Y, Lu Y, Chen W, Fu J, Fan R, Tucker-Kellogg G. In silico experimentation of glioma microenvironment development and anti-tumor therapy. PLoS Comput Biol. 2012;8(2):e1002355. doi:10.1371/journal.pcbi.1002355
26. Pinton L, Masetto E, Vettore M, et al. The immune suppressive microenvironment of human gliomas depends on the accumulation of bone marrow-derived macrophages in the center of the lesion. J Immunother Cancer. 2019;7(1):58. doi:10.1186/s40425-019-0536-x
27. Wu A, Wei J, Kong L-Y, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12(11):1113–1125. doi:10.1093/neuonc/noq082
28. Müller A, Brandenburg S, Turkowski K, Müller S, Vajkoczy P. Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells. Int J Cancer. 2015;137(2):278–288. doi:10.1002/ijc.29379
29. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi:10.1038/ni.1937
30. Wang Y, Lin Y-X, Qiao S-L, et al. Polymeric nanoparticles promote macrophage reversal from M2 to M1 phenotypes in the tumor microenvironment. Biomaterials. 2017;112:153–163. doi:10.1016/j.biomaterials.2016.09.034
31. Dello Russo C, Lisi L, Tentori L, et al. Exploiting microglial functions for the treatment of glioblastoma. Curr Cancer Drug Targets. 2017;17(3):267–281. doi:10.2174/1568009616666160813191240
32. Zhang M, Hutter G, Kahn SA, et al. Anti-CD47 treatment stimulates phagocytosis of glioblastoma by M1 and M2 polarized macrophages and promotes M1 polarized macrophages in vivo. PLoS One. 2016;11(4):e0153550. doi:10.1371/journal.pone.0153550
33. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol. 2014;5:614. doi:10.3389/fimmu.2014.00614
34. Saha D, Martuza RL, Rabkin SD. Macrophage polarization contributes to glioblastoma eradication by combination immunovirotherapy and immune checkpoint blockade. Cancer Cell. 2017;32(2):253–267.e255. doi:10.1016/j.ccell.2017.07.006
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.