Back to Journals » Infection and Drug Resistance » Volume 11
Overproduction of efflux pumps caused reduced susceptibility to carbapenem under consecutive imipenem-selected stress in Acinetobacter baumannii
Authors Zhang Y , Li Z, He X, Ding F, Wu W, Luo Y, Fan B, Cao H
Received 11 September 2017
Accepted for publication 23 December 2017
Published 29 March 2018 Volume 2018:11 Pages 457—467
DOI https://doi.org/10.2147/IDR.S151423
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Yanpeng Zhang,1,2 Zhuocheng Li,2 Xiaolong He,1 Fanglin Ding,2 Weiqing Wu,2 Yong Luo,2 Bing Fan,2 Hong Cao1
1Department of Microbiology, Guangdong Provincial Key Laboratory of Tropical Disease Research, School of Public Health, Southern Medical University, Guangzhou, 510515, China; 2Laboratory Department of the First Affiliated Hospital of Shenzhen University, Shenzhen, 518000, China
Purpose: Acinetobacter baumannii is an important pathogen in the nosocomial infections worldwide. Combining with carbapenemases, efflux pumps and outer membrane proteins (OMPs) have been thought to affect the development of carbapenem resistance in A. baumannii. This study aimed to investigate the contributions of different efflux pumps and OMPs in developing carbapenem resistance in a clinical isolate of A. baumannii and reveal the possible mechanism of overproduction of main efflux pumps.
Patients and methods: In this study, an imipenem-susceptible clinical isolate was identified as A. baumannii and named SZE. Several common carbapenemases were detected by polymerase chain reaction (PCR). Imipenem-selected mutants were selected from SZE by serial subcultivations on Mueller–Hinton agar, and the minimum inhibitory concentration (MIC) was detected. Gene expressions of four families of efflux pumps, five OMPs, and blaOXA-51 were determined by reverse transcription quantitative PCR, and comparisons were made between SZE strain and the imipenem-selected mutants. The adeRS system in SZE and its mutant was sequenced and aligned.
Results: Under consecutive imipenem-selected stress, the MIC to imipenem increased gradually from 0.125 μg/mL to 8 μg/mL. The effect of resistance inducement was almost neutralized when treated with an efflux pump inhibitor. The expression of efflux pumps, adeB, adeG, and adeJ, was increased by 6.9-, 4.0-, and 2.1-fold in mutants, respectively, compared to SZE. A single mutation (G to A) at position 58 was detected in the regulatory adeRS system and possibly upregulated the adeB expression, and then affected the carbapenem resistance in A. baumannii strains.
Conclusion: In conclusion, under consecutive imipenem-selected stress in vitro, A. baumannii strain evolved the ability to reduce susceptibility to a variety of antimicrobials by overproduction of efflux pumps. Especially, the resistance-nodulation-cell division super family and a nucleotide mutant in adeRS regulating system caused the overexpression of adeABC.
Keywords: Acinetobacter baumannii, efflux pump, multidrug resistance, imipenem, outer membrane protein
Introduction
Acinetobacter baumannii is a Gram-negative nonfermentative coccobacillus. In recent years, this opportunistic pathogen has emerged as one of the main causes of hospital-acquired infections, such as ventilator-associated pneumonia, urinary tract infections, bloodstream infections, and surgical wound infections.1 The risk factors associated with A. baumannii infections include invasive medical procedure, mechanical ventilation, immune suppression, burns, and trauma. Additionally, A. baumannii is one of the most frequently isolated clinical pathogens in China.2 Carbapenems, such as imipenem and meropenem, either alone or in combination with other antibiotics are currently the most effective therapeutic options for A. baumannii infections. However, carbapenem-resistant clinical isolates of A. baumannii have notably increased in recent years, and have resulted in a delay in treatment. It is significant to elucidate the molecular mechanism underlying the resistance of A. baumannii to carbapenem for solving the prevailing issue.
A. baumannii has a plastic genome, which contributes to the acquisition and dissemination of multiple resistance mechanisms. These genome plasticity mechanisms include mutations and insertions of mobile elements, such as plasmids, integrons, transposons, and resistant islands.1 Moreover, the overexpression of intrinsic carbapenemase genes, enzymatic degradation, and modification of the target site decreased permeability, and efflux pumps are common possible mechanisms for the acquisition of resistance. In clinical isolates, chromosomally located resistance island and plasmids carrying different antimicrobial resistance determinants have shown an outstanding ability to rapid evolution of resistance when subjected to the pressure of new antimicrobials.3 Multidrug-resistant Acinetobacter spp. can acquire antimicrobial agent resistance genes via class 1 integrons.4,5
Acquired or endogenous carbapenemase activity, together with decreased outer membrane permeability and overproduction of efflux pumps, constitutes the causes of carbapenem resistance in A. baumannii clinical strains.1,6 Commonly, there are two intrinsic β-lactam hydrolyzing enzymes, AmpC and OXA-51-like, in A. baumannii. Yet these two enzymes show only weak hydrolysis to carbapenems.7 Located upstream of the position of blaOXA-51-like gene, the insertion sequence element ISAba1 has been reported to upregulate the expression of the blaOXA-51-like gene, and accordingly conferred resistance to carbapenem antimicrobials in A. baumannii.8
Main carbapenemases, including Ambler Class D enzymes (also known as oxacillinses, such as OXA-23-like, OXA-24-like, and OXA-58-like) and Ambler Class B enzymes (metallo-β-lactamases, such as NDM-1) can hydrolyze carbapenem antimicrobials.9 Production of OXA-23-like enzymes has been regarded as a main reason for carbapenem resistance in A. baumannii in China.10
Alteration of bacterial membrane permeability is another important resistance mechanism. Low expression of outer membrane protein (OMP) reduces porins in carbapenem-resistant strain. Limansky et al found a loss of a 29-kDa protein, named CarO, associated with carbapenem resistance in A. baumannii.11 Other studies have identified that the loss or reduced expression of a variety of OMPs was associated with carbapenem resistance, including Omp33, OmpA, OmpW, and OprD in recent decades.11–15
Recently, research on efflux pumps resistance mechanism in A. baumannii has generated considerable interest. These pumps can extrude a variety of antimicrobial agents and compounds to the external environment.16 Overproduction of efflux pumps can reduce the accumulation of antibiotics, which could be an efficient mechanism for carbapenem resistance. To date, five super families of efflux pumps have been found to be associated with antibiotic resistance in A. baumannii: the resistance-nodulation-cell division (RND) family,16–18 the ATP-binding cassette (ABC) transporters family, the multidrug and toxic compound extrusion (MATE) family,19 the major facilitator super (MFS) family, and the small multidrug resistance (SMR) family.20,21Among these pumps, three members of the RND family, AdeABC, AdeFGH, and AdeIJK, are regarded as the most important for extruding a quite broad range of substrates. The AdeABC pump is the first identified RND-type efflux pump, in which AdeB acts as the antimicrobial transporter, AdeC as the OMP, and AdeA as the periplasmic linking protein. Overproduction of AdeABC confers decreased susceptibility to a variety of antimicrobials, including carbapenems, and it is regarded that other efflux mechanisms also contribute in this process.20 A two-component regulatory system, AdeRS, has been proved to be responsible for the regulation of expression of AdeABC,22 and mutations in AdeRS can result in the overproduction of this pump. AdeL, a LysR-type transcriptional regulator, located upstream from the AdeFGH operon and transcribed in the opposite direction has been found to be responsible for the overproduction of AdeFGH.17 AdeIJK is another RND-type efflux pump and is regulated by AdeN, a TetR-type regulator in A. baumannii.23
As an important clinical antimicrobial, carbapenem might be a potent inducer of multidrug resistance in A. baumannii.24 Thus, continuous treatment with carbapenems in control of infections may generate some carbapenem-selected A. baumannii mutants. Carbapenemases combined with active efflux pumps and decreased permeability of membrane have been illustrated to be main mechanisms of carbapenem resistance. However, under the stress of carbapenem, how a clinical strain developed carbapenem resistance in A. baumannii remains unclear, and few investigations have focused on the efflux pump mechanism as an independent contributor to carbapenem resistance in A. baumannii. In this study, we acquired mutants of a clinical strain under consecutive imipenem-selected stress in vitro. We compared the changes of expressions of efflux pumps and OMPs in the original strain and its carbapenem-selected mutants, and found that the substitution (G to A) at the position 58 in adeRS regulatory system might be the main cause of overexpression of adeABC.
Patients and methods
Strain isolation and identification
A multidrug-resistant Acinetobacter calcoaceticus–A. baumannii complex was selected from 752 clinical isolates collected from 2015 to 2016 at the First Affiliated Hospital of Shenzhen University in China. This strain was isolated from the urine sample of a patient who suffered from a urinary tract infection and was identified as A. calcoaceticus–A. baumannii complex using the Vitek system (BioMerieux, Marcy l’Etoile, France). Then the strain was further identified as A. baumannii using the 42°C growth experiment and the rpoB gene sequence analysis with a previously described method and named as SZE strain.25,26 This study was approved by the Ethics Committee of the First Affiliated Hospital of Shenzhen University.
Detection of carbapenemases
According to Ambler’s classification for β-lactamases, the main carbapenemases, blaNDM-1 (B group), blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like (D group) genes were detected according to previously established polymerase chain reaction (PCR) reaction mixture and thermal conditions.10,27 The detection of ISAba1/blaOXA-51-like gene combination was made referring to the method of Turton et al.8 A. baumannii reference strain ATCC 17978 and three A. baumannii isolates (ab606, ab608, and ab609 [carrying blaOXA-58-like, blaOXA-23-like, and blaOXA-24-like genes, respectively]) were used as the quality control strains for the detection of carbapenemases.
Consecutive selection for imipenem resistance, efflux pump inhibition test, and susceptibility monitoring
Imipenem-selected mutants were selected from SZE strain using a serial subcultivation method on Mueller–Hinton (MH) plate. In the selection of subculture, a common 10 µg containing imipenem disk was placed on the MH plate, and the clone in the margin of the inhibition zone was isolated as the inoculums for the next passage. Due to the diffusion of imipenem from the imipenem-impregnated disk, these clones were in the zone of subinhibitory concentration of imipenem. In this manner, imipenem-selected mutants were selected from SZE strain. Then, imipenem was removed and subcultivation was completed.
To determine the contribution of efflux pumps in SZE strain and its imipenem-selected strains, efflux pump inhibition test was carried out on MH agar plates with and without the efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma-Aldrich Co., St. Louis, MO, USA). The concentration of CCCP used in the test was one-half of the minimum inhibitory concentration (MIC) of CCCP, which was determined by the microdilution method according to the guidelines of the Clinical Laboratory Standards Institute (CLSI). The Kirby–Bauer agar diffusion method (KB method) was used to monitor the susceptibility to imipenem with and without the efflux pump inhibitor.24
Antimicrobial susceptibility testing
The antimicrobial susceptibility of SZE strain and its imipenem-selected strains was determined by broth microdilution method using the following antibiotics with and without the efflux pump inhibitor CCCP. The following antimicrobials were included: β-lactams (ampicillin, ceftazidime, imipenem, meropenem, and ampicillin/sulbactam), aminoglycosides (amikacin, gentamicin, kanamycin, and tobramycin), quinolones (ciprofloxacin and levofloxacin), and tetracyclines (tetracycline).The antimicrobial susceptibility testing of intermediate mutants (G1, G10, G20, G30, G45, and G50) was determined in the presence of CCCP with a concentration one-half of the MIC of CCCP, which was determined by the microdilution method according to the guidelines of the CLSI.
The results were interpreted according to the guidelines of the CLSI. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were used as references for antimicrobial susceptibility testing.
Relative expression of efflux pumps, OMPs, and carbapenemase blaOXA-51-like in SZE (G1) and its imipenem-selected strains (G10, G20, and G45)
Genes of intrinsic carbapenemase blaOXA-51-like, four families of efflux pumps, and five OMPs were selected to compare their expressions in SZE isolate (G1) and its imipenem-selected strains G10, G20, and G45. Expression of three RND family efflux pump members (adeB, adeG, and adeJ) was detected in G10, G20, and G45 intermediate mutants. The genes craA, amvA, and tetB (MFS family), abeM (MATE family), and abeS (SMR family) were detected in G1 and G45 mutants. For detecting the expression of the OMP, the carO, omp33, ompA, ompW, and oprD genes were selected in G1 and G45 mutants. The primers used are listed in Table 1.
  | Table 1 DNA sequence of primers and amplicon size Abbreviations: F, forward; R, reverse. |
RNA isolation was performed using a HiPure Bacterial RNA Kit (Megon Biological Company, Guangzhou, China) according to the manufacturer’s guidelines, and the RNA sample was dissolved in 50 μL RNase-free water. The quality of the RNA sample was determined using the ultraviolet spectrophotometer method. Reverse transcription quantitative PCR (RT-qPCR) was performed using a previously reported method.28 All primers used in the RT-qPCR reactions are listed in Table 1, and the reactions were carried out using a Bestar™ qPCR RT Kit (Xihan Biological Company, Shanghai, China). In a 20 μL reaction mixture, 5 μL cDNA, 10 μL Bestar® SYBR Green qPCR Master Mix (Xihan Biological Company), and 0.2 μM concentration each of forward and reverse primers were included. The reactions were performed with one cycle at 95°C for 2 minutes followed by 45 cycles of 95°C for 10 seconds, 60°C for 34 seconds (fluorescence signal was collected), and 72°C for 30 seconds. In each reaction, the 16S rRNA gene was used as an endogenous control, and all reactions were done in biological triplicate.
Sequencing for regulatory genes of RND-type efflux pumps: adeRS, adeL, and adeN
We investigated sequence information of the regulatory genes adeRS, adeL, and adeN, which act as transcriptional regulators of adeABC, adeFGH, and adeIJK, respectively. Primers were designed according to the published sequence of ATCC 17978. The PCR products were sequenced for BLAST analysis. The primers used are listed in Table 1.
Statistical analysis
Data entry and analyses were processed with Statistical Package for Social Sciences (SPSS Inc., Chicago, IL, USA) version 16.0. Comparison of transcription of resistance genes between SZE strain and its imipenem-selected mutants was carried out by the Student’s t-test. All tests were two-tailed and a p-value ≤0.01 was considered to be statistically significant.
Results
Clinical characteristics
The SZE strain was isolated from the urine sample of a patient who suffered from a urinary tract infection. Bacteria were quantified as 1.05×106 colony-forming units (CFU)/mL in urine specimens, and white blood cell count of 32.76×109/L, C-reaction protein level of 51.38 mg/L, and procalcitonin level of 0.41 ng/mL were detected in the blood sample of the patient.
The SZE strain was identified as A.calcoaceticus–A. baumannii complex by the Vitek system. This strain could grow at a surrounding temperature of 42°C, and was further identified as A. baumannii through the rpoB gene sequence analysis. (GenBank accession number: MF678797) The SZE strain was resistant to nearly all classes of antimicrobial agents, including penicillins, cephalosporins, aminoglycosides, and quinolones. Meanwhile, the SZE strain was susceptible to imipenem and meropenem (Table 2).
Detection of carbapenemase genes
As an intrinsic gene of A. baumannii, the carbapenemase blaOXA-51-like was found in SZE. The other carbapenemases, including the blaNDM-1, blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like genes, were not found in SZE or its imipenem-selected strains. Furthermore, no ISAba1/blaOXA-51-like gene combination was found in these strains.
Consecutive selection for imipenem resistance, efflux pump inhibition test, and susceptibility monitoring
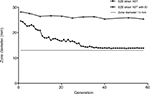
During the resistance inducement process, the diameter of the zone of inhibition declined gradually, and an increased resistance pattern was observed in consecutive subcultivations. As shown in Figure 1, after the first 15 passages, the diameter of the zone of inhibition decreased rapidly, and a constant decline in diameter was observed between the 15th and 20th passages. However, after 30 subcultivations, the decline was nearly invariant. Interestingly, the diameter of the zone of inhibition could not reduce beyond a 13 mm diameter, and resistance in the SZE strain could be induced to the intermediate level but not to be completely resistant to imipenem.
The final imipenem-selected mutant was identified to derive from SZE strain, since the two strains shared the same sequences of rpoB gene.
The MICs of both CCCP in SZE and its imipenem-selected mutants were 128 mg/L, and antimicrobial susceptibility testing was determined in the presence of CCCP at 64 mg/L. Nevertheless, as illustrated in Figure 1, the diameter of the zone of inhibition produced by the imipenem-selected SZE strain was constant with efflux pump inhibitor CCCP.
Antimicrobial susceptibility testing of the SZE strain and its serial subcultivations
A continuous increase in MIC was observed among SZE serial subcultivations for all classes of antimicrobial agents included in this study. And the mutants became resistant to almost all categories of antimicrobials employed in the process of resistance inducement. As shown in Table 2, in the first 10 generations, MIC increased slowly, then increased sharply in the 10th to 20th generations, and stayed unchanged after the 30th generation. In this process, over fourfold of increase of MIC to ampicillin, ceftazidime, imipenem, meropenem, kanamycin, tobramycin, ciprofloxacin, and levofloxacin was observed in the last acquired mutants. The SZE strain was originally susceptible to imipenem and meropenem and the MIC was 0.125 μg/mL, and then an increased MIC at 8 μg/mL of the intermediate level was recorded in the G45 imipenem-selected strain.
Antimicrobial susceptibility testing of the intermediate mutants (G1, G10, G20, G30, G45, and G50) was performed in the presence of CCCP at 64 mg/L. As detailed in Table 2, it was apparent that MICs to all categories of antimicrobials decreased sharply with the CCCP; for instance, the MIC to ampicillin declined from 128 μg/mL to 4 μg/mL in G1. Although the MIC of SZE strain increased from 0.125 μg/mL to 8 μg/mL under the consecutive imipenem-selected stress, the MIC was almost unchanged in the presence of CCCP during the process of inducement. Detailed results are shown in Table 2.
Distribution of efflux pumps and OMP genes in SZE strain (G1) and its imipenem-selected strain (G45)
All efflux pumps and OMP genes could be detected in SZE strain (G1) and its imipenem-induced resistant strain (G45). The efflux pump genes were adeB, adeG, and adeJ (RND family); abeM (MATE family); abeS (SMR family); and cracA, amvA, and tetB (MFS family). The OMP genes were carO, omp33, ompA, ompW, and oprD.
Relative expression of blaOXA-51, efflux pump, and OMP genes in mutants
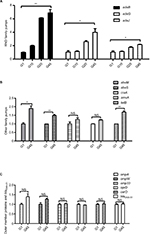
The results of RT-qPCR are shown in Figure 2A–C. As illustrated in Figure 2C, no significant change was found in the relative expression of intrinsic carbapenemase blaOXA-51 in the imipenem-selected strain G45, compared with the original SZE strain.
Changes in the expression of efflux pump families were statistically significant, including the RND family, MATE family, and SMR family. Compared with the original SZE strain G1, the imipenem-selected strain G45 showed a relatively increased expression of the efflux pump gene adeB, adeG, and adeJ (RND family); abeM (MATE family); abeS (SMR family); and tetB (MFS family) (Figure 2A and B). It could be observed that the relative expression of three RND efflux family members, adeB, adeG, and adeJ, increased gradually in the intermediary mutants: by 2-, 1.2-, and 1.1- in G10, then by 6.1-, 2.1-, and 1.8 in G20, then by 6.9-, 4.0-, and 2.1-fold in G45 strains (Figure 2A). The relative expression of efflux pump genes abeM, abeS, and tetB was upregulated by 1.8-, 1.4-, and 1.7-fold, respectively (Figure 2B). Meanwhile, the expression of pump gene of cracA and amvA was unchanged (Figure 2B). On the other hand, the relative expression of the selected OMP genes, including carO, omp33, ompA, ompW, and oprD, did not change significantly in the inducement process (Figure 2C).
Sequence analysis of the regulatory genes for RND pumps: adeRS, adeL, and adeN
The nucleotide sequences of adeR and adeS of both SZE and its imipenem-selected mutant (G45) were acquired for BLAST analysis, and the sequences of adeR of the two strains were submitted to GenBank (GenBank accession number: MF694570 and MF694571). In contrast with the original SZE strain, its imipenem-selected mutant (G45) had a single mutation (G to A) in adeR at the position of nucleotide 58, whereas no nucleotide change was found in adeS. Such replacement would lead to substitution of aspartic acid to asparagine at position 20 of AdeR at the protein level.
Sequence of adeL and adeR, which act as the regulator genes of adeFGH and adeIJK, was sequenced and submitted to GenBank (GenBank accession number: MG266697and MG266698), but no nucleotide change was found in these two genes in the progress of consecutive imipenem selections.
Discussion
A. baumannii has recently posed great challenges in the therapy of nosocomial infections and exhibited resistance to most of the antimicrobial agents. It is necessary to precisely understand the resistance mechanism in clinical isolates of A. baumannii for developing new drugs and raising treatment level to infections of this pathogen. Main resistance mechanisms of A. baumannii include producing carbapenem-hydrolyzing enzymes, decreasing the outer membrane permeability to lessen the entrance of antimicrobial agents, and upregulating efflux pump expression to extrude antimicrobial agents. Frequently, several mechanisms of resistance exist in a multidrug-resistant clinical A. baumannii strain. Few data are available to explain the independent influence of efflux pumps on the acquisition of carbapenem resistance among A. baumannii isolates.
In the present study, we found that imipenem might be a potent inducer for A. baumannii in vitro under the consecutive imipenem-selected stress.24 Upregulation of efflux pumps together with other mechanisms might play an important role for SZE strain in developing carbapenem resistance under the consecutive imipenem-selected stress. A mutation in the adeRS system might be the cause of overproduction of adeABC. A clinical A. calcoaceticus–A. baumannii complex was isolated in our clinical work, and then identified as A. baumannii through the 42°C growth experiments and the rpoB gene sequence analysis, and this strain was named as SZE strain. This isolate was susceptible to both imipenem and meropenem, without carrying any blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, and blaNDM-1 carbapenemase genes. Also, no ISAba1/blaOXA-51-like gene combination was found in this strain. Imipenem-selected mutants were generated from SZE strain by serial subcultivations on MH agar. The imipenem-selected mutants reduced susceptibility to imipenem gradually, with MIC rising from 0.125 μg/mL to 8 μg/mL. The susceptibility to imipenem was monitored in the presence and absence of the efflux pump inhibitor CCCP in this process. Interestingly, the zone of inhibition was almost constant in the presence of CCCP. Although the MIC of SZE strain increased from 0.125 μg/mL to 8 μg/mL under the consecutive imipenem-selected stress, the MIC was almost unchanged in the presence of CCCP during the process. There seemed to be a tendency that increase of resistance during inducement could be neutralized in the presence of inhibitor CCCP, and it could be inferred that overproduction of efflux pumps might be responsible for this increased resistance phenotype.
Correspondingly, the expression of efflux pump genes, especially three members of RND family (adeB, adeG, and adeJ) increased significantly. Other families of efflux pumps also exhibited a relatively increased expression. Under the stress of an antimicrobial agent, a single mutation (G to A) in adeR at nucleotide 58 was detected in imipenem-selected mutant (G45). Such a replacement would lead to substitution of aspartic acid to asparagine at position 20 of AdeR at the protein level.
It has been reported that imipenem is a potent inducer of multidrug resistance in A. baumannii. Kuo et al investigated the progression of multidrug resistance upon exposure to imipenem in A. baumannii strains and found over fourfold increase of MICs to amikacin, cefatazidime, ceftriaxone, ciprofloxacin, meropenem, and imipenem in two A. baumannii reference strains (ATCC 19606 and ATCC17978).24 Correspondingly, they found upregulated mRNA expression of blaOXA-51-like and adeB along with the reduction of adeR and adeS expression only in the ATCC17978 strain. In our study, we found imipenem acted as a potent inducer under the consecutive imipenem-selected stress in vitro, and the MIC to imipenem increased from 0.125 μg/mL to 8 μg/mL, which was consistent with the results of a previous study.24 But in the current study, three members of the RND family of efflux pumps, adeB, adeG, and adeJ, were found to be overexpressed, and a single-nucleotide mutation at the position 58 of the adeRS regulatory system was noticed. Also, the expression of intrinsic blaoxa51 did not change significantly. Under similar condition of imipenem-selected stress, different strains showed different mechanisms, and this indicated that resistance mechanisms might be strain-dependent. Overproduction of RND-type efflux pump has been proved to be associated with resistance to many categories of antimicrobial agents. A mutant with upregulated expression of adeABC, selected on gentamycin gradients, and showing reduced susceptibility to a variety of antimicrobials, including carbapenems (imipenem and meropenem), ceftazidime, cefepime, aminoglycosides, quinolones, and so on, was reported.6 In this study, we obtained a similar result in the imipenem-selected mutants of SZE, and the MICs to β-lactams, aminoglycosides, quinolones, and tetracyclines increased over twofold in this mutant. Several studies reported that the mutation in the regulatory system adeRS resulted in overproduction of adeABC. Higgins et al reported that they isolated a clinical strain of A. baumannii from a patient who suffered from ventilator-associated pneumonia and received combined antimicrobial therapy, including meropenem, ciprofloxacin, amikacin, and cotrimoxazole, and they found a missense mutation in adeR which resulted in >7-fold increase in overproduction of adeB in A. baumannii.29 In our study, we found the same single mutation in vitro under the consecutive imipenem-selected stress, and a replacement of G to A in nucleotide sequence of adeR at nucleotide 58 was detected in imipenem-selected mutant (G45). Such a replacement would lead to a substitution of aspartic acid to asparagine at position 20 of AdeR in protein sequence. AdeR is composed of a signal receiver domain and an effect domain, and this substitution was just on the signal receiver domain in which several phosphorylation sites were located.30 Such a substitution might change the interactions between AdeS and AdeR and then lead to overproduction of adeABC.31 These results suggest that the position 58 was a potent high-frequency mutant site in nucleotide sequence of adeR in clinical isolates which over expressed adeB. Stress induced by different antimicrobials resulted in a similar mutation, and it could be indicated that imipenem might be a potent inducer both in vivo and in vitro.
Through the inducement, the expressions of adeG and adeJ increased by 4.0- and 2.1-fold in mutants, respectively, in our study, and this result was consistent with other recent reports.17,21 AdeIJK was reported to play an important physiological role in intrinsic low-level resistance to various antibiotics among A. baumannii isolates.18,21 As the transcriptional regulator of adeFGH and adeIJK, respectively, the sequences of adeL and adeN remained unchanged in the inducement. This demonstrated that there might be other mechanisms involved in the overexpression of adeFGH and adeIJK.
Parallel to the findings of the present study, the overproduction of several other efflux pumps may induce resistance to imipenem in A. baumannii. Similarly, as a member of the MATE family, abeM was previously reported to extrude aminoglycosides, fluoroquinolones, ethidium bromide, and dyes, and it was overexpressed in this study. The abeS pump, a chromosomally encoded member of the SMR family, was also upregulated during the resistance inducement in the present study.19.
Although no significant change was found in the relative expression of OMP genes including ompA, ompW, omp33, oprD, and carO in this report, it still did not mean that there was no change of expression at the protein level.
Conclusion
In conclusion, under the consecutive imipenem stress in vitro, the A. baumannii SZE strain evolved the ability to reduce susceptibility to a variety of antimicrobials, including carbapenem, through overproduction of efflux pumps, especially the RND super-family efflux pumps. A nucleotide mutant in adeRS regulating system might be the cause of overproduction of adeABC.
Acknowledgment
This project was financially supported by the grant from Shenzhen Science and Technology Innovation Committee, China (Grant No: JCYJ20140414170821183).
Author contributions
HC, YZ, ZL, and XH conceived and designed the experiment; YZ, XH, WW, YL, BF, and ZL performed the experiment; YZ, XH, and ZL analyzed the data; ZL, FD, and HC contributed reagents/materials/analysis tools; YZ, XH, and BF participated in its design and coordination and helped to draft the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work
Disclosure
The authors report no conflicts of interest in this work.
References
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.