Back to Journals » OncoTargets and Therapy » Volume 10
Overexpression of UHRF2 in intrahepatic cholangiocarcinoma and its clinical significance
Authors Peng R, Huang X, Zhang C, Yang X, Xu Y, Bai D
Received 17 August 2017
Accepted for publication 5 November 2017
Published 11 December 2017 Volume 2017:10 Pages 5863—5872
DOI https://doi.org/10.2147/OTT.S149361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Rui Peng,1,2,* Xiaoyong Huang,3,* Chi Zhang,2,* Xuan Yang,3,4,* Yaping Xu,5 Dousheng Bai2
1The Second Affiliated Hospital of Xiangya School of Medicine, Central South University, Hunan, 2Department of Hepatobiliary and Pancreatic Surgery, Subei People Hospital, Clinical Medical College of Yangzhou University, Jiangsu, 3Liver Cancer Institute, Zhongshan Hospital, 4Key Laboratory of Carcinogenesis and Cancer Invasion, Fudan University, Ministry of Education, Shanghai, 5Department of Gastroenterology, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: Ubiquitin-like with PHD and ring finger domains 2 (UHRF2) has been implicated in tumorigenesis. However, its roles in intrahepatic cholangiocarcinoma (ICC) are still unclear. In this study, UHRF2 expression was analyzed in several kinds of cancers by referring to public Oncomine database, and the levels of UHRF2 mRNA and protein were determined in ICC cells and tissues. Then, the roles of UHRF2 in ICC were investigated by UHRF2 interference. Moreover, the relationship between UHRF2 and E-cadherin expression was examined in ICC cells and samples. Finally, the prognostic role of UHRF2 in ICC was analyzed in 139 ICC patients by Cox regression and Kaplan–Meier methods. We found UHRF2 was overexpressed in multiple human cancers, as well as in ICC, and the invasion, migration, proliferation, and antiapoptosis of ICC cells were inhibited by UHRF2 interference. Moreover, the epithelial–mesenchymal transition-related marker E-cadherin was upregulated in ICC cells which was influenced by UHRF2 expression. Clinically, UHRF2 expression was positively associated with microvascular invasion and lymphatic metastasis of ICC, and patients in the UHRF2high group had much lower overall survival and higher recurrence rates than patients in the UHRF2low group. A multivariate analysis revealed that UHRF2 overexpression was a new prognostic marker for ICC. Thus, our results indicated that high level of UHRF2 might be a novel predictor for the prognosis of ICC.
Keywords: ICC, UHRF2, E-cadherin, prognosis, biomarker
Introduction
Intrahepatic cholangiocarcinoma (ICC), a rare malignant tumor derived from the intrahepatic and extrahepatic biliary tract, has a poor prognosis and a high incidence.1 Despite the development of novel diagnostic tools and clinical screenings in recent years, the resection rate of ICC is still low and variable (18%–70%) due to a majority of patients being diagnosed at an advanced stage.2 Moreover, most of the post-resection patients show recurrence (up to 75%) in the remnant liver after the curative treatment.3 Thus, it is important to reveal the underlying mechanism for ICC development.
Ubiquitin-like with PHD and ring finger domains 2 (UHRF2) is a member of the UHRF family proteins, which includes an ubiquitin-like domain, a plant homeodomain, a RING finger domain, and a SET and RING finger-associated (SRA) domain. As a ubiquitin E3 Ligase, UHRF2 was initially identified as a cell cycle regulator by its interaction with the inactive CDK2–cyclin E complex.4 Then, UHRF2 was demonstrated to bind to H3K9me2/me3-containing peptides, and interact chromatin-mediated genes with SRA.5–7 Recently, it has been further found to function as a key epigenetic regulator by interacting with DNA and histone methyltransferases.8,9 Recently, the roles of UHRF2 in cancer have emerged; for example, UHRF2 was found to induce the apoptosis of tumor cells by regulating E2F110,11 and to reduce the expression of cell cycle-related proteins in lung cancer and malignant glioma, which indicated that UHRF2 functioned as a tumor suppressor.12,13 More recently, UHRF2 was uncovered to be a promoter of tumor development by inhibiting the expression of tumor suppressor genes, such as p16, p21, and p27, through its action on DNA methylation and histone modification in breast cancer.14 Furthermore, high level of UHRF2 was revealed to induce ERK1/2 activity via transcriptional deregulation, and to be negatively associated with the patients’ overall survival (OS) in colon cancer, which indicated that UHRF2 might be an oncogene.15,16 The above results indicate that UHRF2 is a multistructural protein and has a wide range of functions, and its function in tumors depends on the tumor type and cellular context.
Epithelial–mesenchymal transition (EMT) is a multistep biologic process in which the epithelial cells transform to mesenchymal phenotype cells. Now, EMT is deemed as an essential element in tumor metastasis of several epithelial malignancies.17 E-cadherin, an important molecule in cell EMT process, localizes at regions of cell-cell contact, which is down-regulation during loss of epithelial cells polarity and gain of mesenchyme cells migratory properties.18 So, the expression of E-cadherin is a marker to estimate tumor invasion and metastasis.19,20 Recently, UHRF2 was demonstrated by proteomics analysis to promote tumor progression by inducing cell EMT.21 However, the relationship between UHRF2 and E-cadherin expression in most tumors is still unclear, which needs to be further elaborated.
This study would investigate the expression and functions of UHRF2 in ICC and assess the relationship between UHRF2 and E-cadherin expression in ICC cells and tissues. In addition, the clinical implication of UHRF2 expression in ICC was analyzed.
Patients and methods
Patients and samples
Eighteen pairs of frozen tissues from ICC patients were obtained from the tissues center of Zhongshan Hospital, and the ICC tissues were acquired from 139 patients who underwent curative surgical resection at the Department of Liver Surgery of Zhongshan Hospital, Fudan University, between February 2003 and November 2010. Both tumor and peritumor samples were obtained from each patient. Complete removal of tumor nodules including peripheral lymph node, hepatoduodenal ligament, and hepatic bile duct is referred to as curative resection. The histopathologic diagnosis of ICC was according to the World Health Organization standard. The Child Pugh score system was used to assess liver function. The tumor stage of the patients was determined according to the TNM sixth edition of the Union for International Cancer Control. Follow-up data were obtained until February 2015. The median survival time was 18 months (range 4–120 months). This study was approved by the Zhongshan Hospital Research Ethics Committee. All the participants provided an informed consent. All procedures in this study were performed according to the ethical standards of the institutional and/or national research committee and in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Tissue microarrays (TMAs) and immunohistochemistry
The construction of TMAs and immunohistochemistry were performed as previously described.22 Rabbit anti-human UHRF2 monoclonal antibody (1:1,000; Wuhan Boster Biological Technology, Ltd., Wuhan, People’s Republic of China) and rabbit anti-human E-cadherin antibody (1:100; Cell Signaling Technology, Danvers, MA, USA) were utilized to detect the levels of UHRF2 and E-cadherin. The density level of strong, moderate, and weak staining was defined as in previous studies.22–24 The assessment was performed blindly by two pathologists.
Cell lines and cell culture
Two ICC cell lines, RBE and QBC939, were provided by the Chinese Academy of Sciences Shanghai branch cell bank (Shanghai, People’s Republic of China). These two cell lines were cultured in RPMI-1640 supplemented with 10% fetal bovine serum, streptomycin, and penicillin at 37°C in a humidified incubator under 5% CO2.
Small interfering RNA
Two different siRNA sequences were purchased from Shanghai Genomeditech Company (Shanghai, People’s Republic of China). The target sequences are as follows: siRNA1: GTCAGAGTGATACGCAGTT; siRNA2: GGAGATTATGTCTACGTTT. The siRNA and the negative control were transfected using lipofectamine™ 2000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the protocols. The efficiency of silence was determined after 72 hours of transfection.
Cell proliferation, migration, and invasion assays, flow cytometry
Cell proliferation was estimated by cell counting kit-8 assay (Yeason, Shanghai, People’s Republic of China). Migration ability was analyzed by wound healing assay. Invasion was evaluated by transwell assay. Cell apoptosis was detected by flow cytometry (Becton Dickinson). Each protocol has been published earlier.23–25
Western blot and quantitative real-time polymerase chain reaction (qRT-PCR)
Western blot and immunofluorescence were performed as in our early studies.22–24 UHRF2 (1:1,000; Boster) and E-cadherin mABs (1:1,000; Abcam, Cambridge, MA, USA) were used to detect the expression of UHRF2 and E-cadherin (1:1,000) in three ICC cell lines. Tubulin (1:1,000; Cell Signaling Technology) was used as the internal control.
The primers used for PCR are as follows: UHRF2: 5′-GGACCTTCCAATCAGCCATC-3′ (forward), 5′-GACATCTCTGGCATCCACCA-3′ (reverse); E-cadherin: 5′-GAACGCATTGCCAC ATACAC-3′ (forward), 5′-GAATTCGGGCTTGTTGTCAT-3′ (reverse); tubulin: 5′-CTGCCTGTT GTACCGTGGTG-3′ (forward), 5′-ATGCCAACCTTGAAGCCAGT-3′ (reverse).
Immunofluorescence assays
Cells were treated with 0.1% Triton X-100 for 30 minutes at room temperature. After washing with PBS and blocking with 10% bovine serum albumin for 1 hour, cells were incubated with primary antibody overnight at 4°C. Successively, cells were washed with PBS and treated with secondary antibody for 2 hours, and the cell nuclei were stained by 4′,6-diamidino-2-phenylindole. Finally, the slides were photographed with a fluorescence microscope.
Statistical analysis
Quantitative data were analyzed using Student’s t-test between two groups. Categorical data were analyzed using the χ2 or Fisher’s exact tests. The Kaplan–Meier method was used to determine survival probability, and differences were assessed by the log-rank test. Cox analysis was used to determine independent prognostic factors. A P-value of <0.05 was considered statistically significant. All analyses were performed using SPSS 21.0 software (IBM Corporation, Armonk, NY, USA).
Results
UHRF2 overexpressed in human cancers
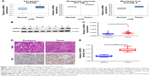
Firstly, we tried to analyze the expression of UHRF2 in multiple kinds of cancers by referring the Oncomine database. As shown in Figure 1A, the mRNA levels of UHRF2 were increased in the tumor tissues compared with normal tissues, including gastric cancer,26 colon cancer,27 and liver cancer.28
Then four pairs of tissues from ICC patients were randomly chosen to examine UHRF2 protein expression, and we found that UHRF2 expression was much higher in the tumor tissues than that in the corresponding peritumor tissues (Figure 1B). Similarly, UHRF2 mRNA was higher in the tumor tissues than in matched adjacent tissues (P=0.028; Figure 1C). Immunohistochemistry results suggested that the staining intensity of UHRF2 in ICC tissues was characteristically stronger than that in their peritumor tissues (P<0.001; Figure 1D and E). The above results indicated that the UHRF2 level was elevated in ICC tissues.
UHRF2 expression was related to malignant features of ICC
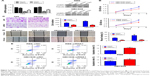
UHRF2 expression was knocked down in QBC939 and RBE ICC cell lines by siRNA (Figure 2A). Then we chose the most silenced siRNA to investigate whether UHRF2 is related to tumor proliferation, aggressiveness, and apoptosis. The results of cell counting kit-8 assays strongly indicated that the ICC cell proliferation was inhibited after UHRF2 interference on the 4th day (Figure 2B). The transwell assay displayed that downregulation of UHRF2 suppressed the invasion of ICC cells (Figure 2C), which was in line with the migration assay (Figure 2D). Moreover, the assessment of cell apoptosis revealed that UHRF2 interference increased ICC cell apoptosis (Figure 2E).
UHRF2 was negatively associated with E-cadherin expression in ICC
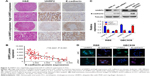
In ICC tissues, UHRF2 and E-cadherin expression exhibited heterogeneity. UHRF2 was restricted to the cytoplasm of ICC cells, while E-cadherin was located at the membranes of the cancer cells. Representative samples are presented in Figure 3A (UHRF2high/E-cadherinlow; UHRF2low/E-cadherinhigh).
In a previous study, reduction of E-cadherin was reported to correlate with the aggressive feature of tumor.20 Here, we found that E-cadherin staining intensity was weak in 63 tissues and UHRF2 staining was negatively related to E-cadherin expression in ICC tissues (r2=0.4221, P<0.001; Figure 3B). In RBE and QBC939 cell lines, we also found the expressions of UHRF2 and E-cadherin were opposite (Figure 3C and D).
High level of UHRF2 was correlated with the poor prognosis of ICC patients
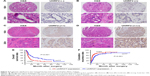
According to the UHRF2 expression, all patients in the TMAs were distributed to two subgroups, including UHRF2low (absence and weak staining) and UHRF2high (moderate and strong staining) groups (Figure 4A–D). To determine the clinical significance of UHRF2 expression in ICC patients, survival curves were created, and we found that overexpressed UHRF2 was definitely correlated with the poor prognosis of ICC patients enrolled in this study. The OS rates of ICC patients with UHRF2low were obviously higher than those of patients with UHRF2high (P<0.001; Figure 4E). The 2- and 5-year OS rates in the UHRF2high group were substantially decreased compared with that in the UHRF2low group (22.6% vs 44.2% and 17.7% vs 26.0%, respectively). The cumulative recurrence rates of ICC patients with UHRF2low were much lower than those of patients with UHRF2high (P<0.001; Figure 4F). The 2- and 5-year cumulative recurrence rates in the UHRF2low group were clearly elevated compared with those of the UHRF2high group (80.6% vs 63.6% and 81.0% vs 75.3%, respectively).
The relationship between the clinicopathologic characteristics of ICC and UHRF2 levels was analyzed. Two factors, microvascular invasion (P=0.028) and lymph node metastasis (P=0.009), were significantly associated with UHRF2 (Table 1). In univariate analysis, tumor size, multiple tumor, embolus, tumor differentiation, lymphatic metastasis, and UHRF2 expression were predictors for OS and cumulative recurrence of ICC patients (Table 2). In multivariate Cox proportional hazards model, expression of UHRF2 was an independent prognostic factor for OS (P<0.001) and cumulative recurrence of ICC patients (P=0.034; Table 2).
Discussion
We found in this study that UHRF2 was overexpressed in ICC tissues, and interference of UHRF2 depressed cancer cell proliferation, invasion, and metastasis and promoted apoptosis. Moreover, we found that the expression of UHRF2 negatively correlated with E-cadherin, loss of which has been linked to poor prognosis of many types of cancer, including ICC. Additionally, high level of UHRF2 was found to be associated with the malignant phenotypes of ICC, including microvascular invasion and lymph nodes metastasis. Finally and importantly, patients with a high level of UHRF2 had poorer OS and high cumulative recurrence rates. These findings indicated that UHRF2 might be a new therapeutic target and a unique prognostic factor.
As an E3 ligase, the understanding of UHRF2 function in cancer experienced a complicated process. Firstly, it was reported to work as a tumor suppressor gene by not only promoting the propagation phase, but also significantly inhibiting the G0/G1 phase.7 For example, cancer outlier profile analysis indicated that UHRF2 was down-regulated in several malignant tumors, such as brain and central nervous system cancer.12,13 Then, it was reported to regulate the posttranslational modification of histones, including H3K9ac and H3K14ac, to induce initiation and progression of tumor.9,10 Meanwhile, UHRF2 was found to interact with hepatitis B core and downregulate acetylation of hepatitis B virus ccc-DNA. A further study has reported its N-domain played an important role in protein degradation or gene transcription.29 Recently, overexpression of UHRF2 has been reported in several types of tumors, including breast cancer, lung cancer, brain cancer, and colon cancer. Specially, UHRF2 interacts with EMT-transcription factors, including ZEB1/2 and SNAI1/2, in gastric cancer, which indicates that UHRF2 could silence the expression of E-cadherin through directly binding the promoter region of E-cadherin gene.21 All of these suggested UHRF2 overexpression promoted tumor progression and might be a potential hallmark for diagnosis and a therapeutic target. Herein, we have presented the expression and functions of UHRF2 in ICC cells and tissues, particularly the clinical implication of UHRF2 in ICC patients, which indicate that UHRF2 overexpression served as a promoter of ICC.
EMT is a critical mechanism for epithelial tumor development.30 EMT is an early factor in metastasis of tumor because mesenchymal phenotype activation and repression of the epithelial phenotype are necessary for the cell to detach from the tumor mass.31,32 The membrane surface E-cadherin, one of the EMT-related proteins, has a fundamental role in EMT. E-cadherin can be repressed by several EMT-inducing transcription factors, including SNAIL1/2, ZEB1/2 and TWIST. Recently, UHRF2 was demonstrated to regulate the functions of these transcription factors.21,31 Thus, UHRF2 may indirectly regulate E-cadherin expression. In this study, we showed that the expression of UHRF2 was negatively associated with E-cadherin expression in ICC tissues. Moreover, the interference of UHRF2 in ICC cells upregulated E-cadherin in the ICC cells. The above results lend support to the notion that UHRF2 does make a substantial contribution to ICC progression by regulating E-cadherin.
Although the precise role of interaction of UHRF2 and E-cadherin in tumor development in vitro and in vivo and other EMT markers needs to be further investigated, our results are important, and detection of UHRF2 may be used as a predictor to find the prognosis in noninvasive and nonmetastatic ICC.
Acknowledgments
This study was funded by the six talent peaks project in Jiangsu Province (2014-WSW-076), the fourth phase of the “333 project” in Jiangsu Province (BRA2015188), the National Natural Science Foundation of China (81502526, 81702861), and the Science and Technology Project of Guangdong Province (2014A030310007).
Disclosure
This is an original work by all the authors and no previous presentations, reports, or publications contain any material that appears in the article. The authors report no conflicts of interest in this work.
References
Stephenson B, Shimwell N, Humphreys E, et al. Quantitative assessment of the cell surface proteome to identify novel therapeutic targets in cholangiocarcinoma. Lancet. 2015;385(Suppl 1):S94. | ||
Raggi C, Invernizzi P, Andersen JB. Impact of microenvironment and stem-like plasticity in cholangiocarcinoma: molecular networks and biological concepts. J Hepatol. 2015;62(1):198–207. | ||
Klempnauer J, Ridder GJ, Werner M, Weimann A, Pichlmayr R. What constitutes long-term survival after surgery for hilar cholangiocarcinoma? Cancer. 1997;79(1):26–34. | ||
Li Y, Mori T, Hata H, Homma Y, Kochi H. NIRF induces G1 arrest and associates with Cdk2. Biochem Biophys Res Commun. 2004;319(2):464–468. | ||
Mori T, Li Y, Hata H, Kochi H. NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 2004;557(1–3):209–214. | ||
Zhang J, Gao Q, Li P, et al. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011;21(12):1723–1739. | ||
Mori T, Li Y, Hata H, Ono K, Kochi H. NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem Biophys Res Commun. 2002;296(3):530–536. | ||
Liu Y, Zhang B, Kuang H, Korakavi G, Lu LY, Yu X. Zinc finger protein 618 regulates the function of UHRF2 (Ubiquitin-like with PHD and ring finger domains 2) as a specific 5-hydroxymethylcytosine reader. J Biol Chem. 2016;291(26):13679–13688. | ||
Zeng S, Wang Y, Zhang T, Bai L, Wang Y, Duan C. E3 ligase UHRF2 stabilizes the acetyltransferase TIP60 and regulates H3K9ac and H3K14ac via RING finger domain. Protein Cell. 2017;8(3):202–218. | ||
Lu H, Hallstrom TC. The nuclear protein UHRF2 is a direct target of the transcription factor E2F1 in the induction of apoptosis. J Biol Chem. 2013;288(33):23833–23843. | ||
Mori T, Ikeda DD, Fukushima T, Takenoshita S, Kochi H. NIRF constitutes a nodal point in the cell cycle network and is a candidate tumor suppressor. Cell Cycle. 2011;10(19):3284–3299. | ||
He X, Duan C, Chen J, et al. Let-7a elevates p21(WAF1) levels by targeting of NIRF and suppresses the growth of A549 lung cancer cells. FEBS Lett. 2009;583(21):3501–3507. | ||
Wu TF, Zhang W, Su ZP, et al. UHRF2 mRNA expression is low in malignant glioma but silencing inhibits the growth of U251 glioma cells in vitro. Asian Pac J Cancer Prev. 2012;13(10):5137–5142. | ||
Wu J, Liu S, Liu G, et al. Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene. 2012;31(3):333–341. | ||
Reinhardt P, Schmid B, Burbulla LF, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12(3):354–367. | ||
Lu S, Yan D, Wu Z, et al. Ubiquitin-like with PHD and ring finger domains 2 is a predictor of survival and a potential therapeutic target in colon cancer. Oncol Rep. 2014;31(4):1802–1810. | ||
Ke AW, Shi GM, Zhou J, et al. CD151 amplifies signaling by integrin alpha6beta1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology. 2011;140(5):1629–1641 e15. | ||
Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62(6):1613–1618. | ||
Nitta T, Mitsuhashi T, Hatanaka Y, et al. Prognostic significance of epithelial-mesenchymal transition-related markers in extrahepatic cholangiocarcinoma: comprehensive immunohistochemical study using a tissue microarray. Br J Cancer. 2014;111(7):1363–1372. | ||
van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14(2):121–134. | ||
Lai M, Liang L, Chen J, et al. Multidimensional proteomics reveals a role of UHRF2 in the regulation of epithelial-mesenchymal transition (EMT). Mol Cell Proteomics. 2016;15(7):2263–2278. | ||
Ke AW, Shi GM, Zhou J, et al. Role of overexpression of CD151 and/or c-Met in predicting prognosis of hepatocellular carcinoma. Hepatology. 2009;49(2):491–503. | ||
Zhang C, Bai DS, Huang XY, et al. Prognostic significance of Capn4 overexpression in intrahepatic cholangiocarcinoma. PLoS One. 2013;8(1):e54619. | ||
Zhang C, Liu LX, Dong ZR, et al. Up-regulation of 14-3-3zeta expression in intrahepatic cholangiocarcinoma and its clinical implications. Tumour Biol. 2015;36(3):1781–1789. | ||
Huang XY, Ke AW, Shi GM, et al. AlphaB-crystallin complexes with 14-3-3zeta to induce epithelial-mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology. 2013;57(6):2235–2247. | ||
Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–1857. | ||
Skrzypczak M, Goryca K, Rubel T, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5(10):e13091. | ||
Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. | ||
Qian G, Hu B, Zhou D, Xuan Y, Bai L, Duan C. NIRF, a novel ubiquitin ligase, inhibits hepatitis B virus replication through effect on HBV core protein and H3 histones. DNA Cell Biol. 2015;34(5):327–332. | ||
Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39(3):305–318. | ||
Vaquero J, Guedj N, Claperon A, Nguyen Ho-Bouldoires TH, Paradis V, Fouassier L. Epithelial-mesenchymal transition in cholangiocarcinoma: from clinical evidence to regulatory networks. J Hepatol. 2017;66(2):424–441. | ||
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.