Back to Journals » Cancer Management and Research » Volume 13
Overexpression of the lncRNA AC012213.3 Promotes Proliferation, Migration and Invasion of Breast Cancer via RAD54B/PI3K/AKT Axis and is Associated with Worse Patient Prognosis
Received 28 May 2021
Accepted for publication 20 August 2021
Published 16 September 2021 Volume 2021:13 Pages 7213—7223
DOI https://doi.org/10.2147/CMAR.S322195
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Hongmei Zhong,1– 4 Guilin Zeng,2– 4 Lang He2– 4
1Chengdu University of Traditional Chinese Medicine, Chengdu, 610075, Sichuan Province, People’s Republic of China; 2Department of Oncology, The Fifth People’s Hospital of Chengdu, Chengdu, 611130, Sichuan Province, People’s Republic of China; 3The Fifth People’s Hospital Affiliated to Chengdu University of Traditional Chinese Medicine, Chengdu, 611130, Sichuan Province, People’s Republic of China; 4Chengdu Cancer Prevention and Treatment Institute, Chengdu, 611130, Sichuan Province, People’s Republic of China
Correspondence: Lang He
Department of Oncology, The Fifth People’s Hospital of Chengdu, The Fifth People’s Hospital Affiliated to Chengdu University of Traditional Chinese Medicine, Chengdu Cancer Prevention and Treatment Institute, Chengdu, 611130, Sichuan Province, People’s Republic of China
Tel +86 15182457840
Email [email protected]
Background: Long noncoding RNA (LncRNA) has been shown to mediate the development of human malignancies. However, data on its role in breast cancer remains scant. We aimed to evaluate the prognostic potential of lncRNAs in breast cancer.
Methods: We downloaded data on breast cancer patients from The Cancer Genome Atlas (TCGA) database. Tissues were obtained from The Fifth People’s Hospital of Chengdu. We then used the DESeq2 package to profile the expression of the lncRNAs between the patients and normal samples. Besides, we performed prognosis and survival analysis using survival tools in R package. We then assayed the role of the differentially expressed lncRNAs, AC012213.3 (ENSG00000266289), in cancer cell lines. Quantitative real-time PCR and Western blot analysis were performed to evaluate the expression of the gene in the cell lines and then assessed its role in the progression of breast cancer using cell proliferation (CCK8 and colony formation assays), migration, invasion (transwell and wound-healing assays) and apoptotic (flow cytometry) assays.
Results: Our data showed high expression of lncRNA AC012213.3 in breast cancer tissues and cell lines. The high expression of the AC012213.3 was associated with the worse prognosis and clinical features. Besides, in vitro assays demonstrated that downregulation of AC012213.3 suppresses the proliferation and invasion of breast cancer cells. Further analysis showed that RAD54B is a downstream AC012213.3 target gene and was upregulated in breast cancer. Interestingly, RAD54B expression was associated with shorter survival in breast cancer. In addition, AC012213.3 was shown to facilitate breast cancer progression through the RAD54B/PI3K/AKT axis.
Conclusion: Taken together, our data demonstrated that lncRNA AC012213.3 is upregulated in breast cancer and could enhance breast cancer progression through RAD54B/PI3K/AKT axis.
Keywords: lncRNA, AC012213.3, breast cancer, PI3K
Introduction
Breast cancer is the most prevalent malignant tumor in women and affects about 226,000 women annually.1 Although breast cancer mortality rate dropped by nearly 40% from 1989 to 2016, the five-year survival rate of patients with distant metastasis remains only 27%.1 As a highly heterogeneous disease, the pathogenesis, clinical manifestation and prognosis of breast cancer vary among individuals.2 Clinical diagnostic tools such as tumor size, lymph node status, age, pathological grade, hormone receptors and human epidermal growth factor receptor 2 (HER2) could indicate prognosis of breast cancer patients. Over the past decade, surgery, radiation therapy, chemotherapy and targeted therapy have been the primary treatment options for breast cancer.3 Despite the advances in breast cancer treatment, there is high recurrence rate and low 5-year survival rate in patients with cancer metastasis. It is, therefore, essential to explore more effective therapeutic targets.
Long noncoding RNAs (lncRNAs) are a family of RNA lacking protein‐coding transcripts longer than 200 nucleotides.4 The lncRNA plays major roles in gene regulation such as chromatin modification, mRNA biogenesis, and protein signaling.5 Previous data has shown that lncRNAs could act a potential target for therapeutic intervention in breast cancer.6–9 For instance, a complex between lncRNA LINC02273 and hnRNPL facilitates cancer metastasis,10 and that lncRNA LINC00908 could encoded ASRPS, a small regulatory peptide of STAT3, whose downregulation blocks angiogenesis in triple-negative breast cancer.11 Besides, overexpression of lncRNA HOTAIR has been shown to promote cell progression and conferred tamoxifen resistance in breast cancer.12 Therefore, it is feasible to speculate that lncRNAs might be playing essential roles in breast cancer.
Here, we analysed data from TCGA database and identified AC012213.3, a lncRNA that is highly expressed in both breast cancer tissues and cell lines. Knockdown of the AC012213.3 could significantly inhibit the proliferation and invasion of breast cancer cells. Besides, the data showed that AC012213.3 might act as an oncogene by modulating the RAD54B/PI3K/AKT axis.
Materials and Methods
Bioinformatics Analysis
The breast cancer patient data was obtained from the TCGA database (https://portal.gdc.cancer.gov/). The data included transcriptional profiling and clinical information of a total of 113 normal samples and 1109 cancer samples. The transcriptional profiling files were acquired, annotated and then gene expression profiling was performed. The “Count” data were normalized using the DESeq2 package in R environment and the code used has been uploaded in the Figshare website (https://figshare.com/articles/dataset/Count_data_and_normalization_code/15177633). The normalized count data were used for differentially expressed analysis and then log2 transformed for further analysis. Protein coding genes and the lncRNAs were distinguished according to the biotype information in the reference file (GRCh38.gtf). Survival package in R was used to analyse the patient prognosis while the DESeq2 package was used to perform differential analysis. High (top 50%) and low (bottom 50%) expression groups were divided according to the median expression of specific value.
Tissues and Cells
Eleven paired breast cancer tissues and corresponding paracancerous tissue were obtained from The Fifth People’s Hospital of Chengdu. All the patients gave a signed informed consent and the study protocol was approved the ethics committee of the The Fifth People’s Hospital of Chengdu prior to study commencement. Four breast cancer cell lines (MCF-7, T47D, MDA-MB-231 and MDA-MB-469) and one normal human breast epithelial cell line (MDA-kb2) were purchased the Cell Bank of China Academy of Sciences (Shanghai, China). The cell lines were cultured and maintained at 37°C in 5% CO2.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen Corporation, Carlsbad, CA, USA) cDNA was synthesized using QuantiTect Reverse Transcription Kit (TaKaRa, Dalian, China). The SYBR Green system (miScript SYBR Green PCR kit, Qiagen) was used to amplify and detect the amplicons. The primers used were as follows: RAD54B, forward primer: 5ʹ-GCCAAACACTGATGATTTGTGG-3ʹ; reverse primer: 5ʹ-CCTGAGAAGAATGCGAGATAGC-3ʹ; AC012213.3, forward primer: 5ʹ-GGGCATTACTGGATTGGT-3ʹ; reverse primer: 5ʹ-TGAAGTGGGTGATGTGGT-3ʹ; GAPDH, forward primer: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ, reverse primer: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ.
Western Blot
The cells were cultured to 100% confluency and then proteins were extracted using a total protein extraction kit (P0033, Beyotime, China). The protein samples were mixed with loading buffer and then denatured by heating10 min). The samples were resolved in a SDS/PAGE and then transferred into membrane. The membrane was incubated with primary antibodies (anti-AKT, 1:1000; anti-p-AKT, 1:2000, anti-CyclinD1, 1:500, anti-β-Actin, 1:1000; Proteintech) overnight at 4°C. The blots were washed in 1X PBS and then incubated with a secondary antibody for 2 h at room temperature.
Plasmid Construction and Cell Transfection
AC012213.3 knockdown or RAD54B overexpression were performed using plasmids synthesized by Synthgene (China). The cells were seeded into 80–90% density in a six-well plate and then transfected with 200 ng/μL of the plasmid using Lipofectamine 3000 (Thermo Fisher, USA). Specific shRNA sequence used were: siRNA1: 5ʹ-GCCCAGAATTTACTTAACT-3ʹ, siRNA2: 5ʹ-GGTGAAATCTCCCATTTCT-3ʹ, siRNA3: 5ʹ-CCATGCTCATATTGTACAT-3ʹ.
Colony Formation Assay
The cells were resuspended in 1 mL serum-free medium and then inoculated into a 6-well plate at a concentration of 500 cells per well. 2 mL of the medium with 10% fetal bovine serum (FBS) was added into each well and then replaced on the 5th day. After 12 days, the cells were fixed with 4% formaldehyde and then stained with crystal violet.
Cell Quantification Assay: Cell Counting Kit-8 (CCK8)
The cells were resuspended and then inoculated into a 24-well plate at a concentration of 2×103 cells per well. We observed the cell at 0h, 24h, 48h and 72h. The cells were mixed with the CCK8 reagent and then cultured for 1.5h more before measurement of absorbance.
Transwell Assay
The cell invasion and migration ability was assessed by transwell assay. The transwell chamber was placed in a 24-well plate and then the lower layer of the chamber was filled with 600 µL of medium containing 20% FBS. In parallel, 200 µL serum-free medium with 2×104 cells was added into the upper layer. After 24h, the cells were fixed with formaldehyde. The cells on the lower chamber were stained with crystal violet and then observed under a microscope.
Wound-Healing Assay
The cells were seeded into a six-well plate and cultured to 90–100% confluency. A 200 µL pipette tip was used to perform a vertical or horizontal scratch. 0h and 24h were set as individual observation nodes.
Apoptosis Assay
We employed flow cytometry to evaluate cell apoptosis. Briefly, cells were resuspended into binding buffer and stained with annexin V‐FITC reagent. We then used BD Accuri C6 Plus (BD Biosciences) to analyze the stained cells. The results were processed in FlowJo software.
Statistical Analysis
SPSS, R, Image J and GraphPad Prism 8 softwares were used to perform visualization and statistical analyses. All the experiments were repeated at least three times. Statistical significance between different groups was evaluated using a Students t-test.
Results
AC012213.3 is Upregulated in Breast Cancer and Correlate with Poor Clinical Features
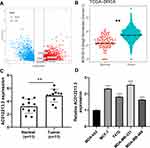
Our analysis showed that there were a total of 3169 differentially expressed lncRNAs between the tumor and normal tissues. 1712 were upregulated while 1457 were downregulated lncRNAs (Figure 1A). We then employed univariate cox analysis to screen prognosis-related lncRNAs in the upregulated lncRNAs. The analysis showed that AC012213.3 was the most significantly upregulated lncRNA in both TCGA and our paired breast cancer tissue (Figure 1B and C, Table S1; HR = 1.16). Similarly, data from cell lines also indicated a remarkably higher expression of AC012213.3 in breast cancer cells compared with normal cells (Figure 1D). In addition, patients with high AC012213.3 expression were shown to have worse clinical features, especially in clinical stage, T and N classification (Figure 2A–D). Furthermore, profiling of the AC012213.3 expression in different immunochemistry subtypes of breast cancer showed that triple-negative breast cancer had the lowest AC012213.3 expression compared with other subtypes. The HER2+HR+ breast cancer had higher AC012213.3 expression compared to the HER2-HR+ breast cancer (Figure 2E). Moreover, the Kaplan-Meier survival curve analysis showed that AC012213.3 was associated with short overall survival of the patients (Figure 2F).
AC012213.3 Promotes Cell Proliferation and Inhibits Cell Apoptosis in Breast Cancer
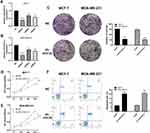
To evaluate the biological roles of AC012213.3 in breast cancer, we performed a series of cell phenotype assays. qRT-PCR showed successful downregulation of AC012213.3 by the siRNA1 sequence in both MCF-7 and MDA-MB-231 cells (Figure 3A and B). Data from the colony formation and the CCK8 assays demonstrated that the suppressed AC012213.3 expression could significantly inhibit the proliferation ability of the breast cancer cells (Figure 3C and D). Furthermore, we showed that the downregulation of AC012213.3 expression could enhance apoptosis (Figure 3D).
AC012213.3 Facilitates Cell Motility in Breast Cancer
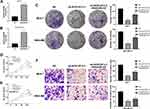
MCF-7 and MDA-MB-231 were selected for the further experiments for their highest AC012213.3 expression. To assess the metastatic ability of cancer cells, we explored the role of AC012213.3 in cell motility. Findings from the Transwell assay demonstrated that the MCF-7 and MDA-MB-231 cell lines bearing a downregulated AC012213.3 had lower invasion and migration activity (Figure 4A). Similarly, the wound-healing assay showed that the suppression of AC012213.3 could significantly hamper the movement ability of breast cancer cells (Figure 4B).
RAD54B Expression Correlates with AC012213.3 and is Linked to Worse Clinical Parameters
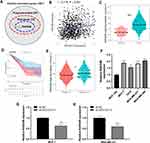
Based on the expression profile from TCGA database, we screened the mRNA which were positively correlated with AC012213.3 (|R| > 0.2 and P < 0.05). A total of 22 genes were shown to be tightly associated with patient prognosis (Figure 5A). RAD54B was among the screened genes, and had been reported to promote the progression of breast cancer cells.13 Further analysis showed significantly positive correlation between RAD54B and AC012213.3 (Figure 5B; R = 0.176, P < 0.001). Besides, there was upregulation of the RAD54B mRNA in the breast tumor tissues compared with the normal tissues (Figure 5C). In addition, Kaplan–Meier survival curves demonstrated that patients with high RAD54B expression had shorter overall survival (Figure 5D; P < 0.001), and were mainly of advanced cancer stages (Stage III–IV) (Figure 5E). Similarly, breast cancer cell lines showed a high expression of RAD54B mRNA (Figure 5F). Besides, in the AC012213.3 knockdown cells, there was increased in the expression of RAD54B compared with the control group, thus the AC012213.3 could positively regulate RAD54B expression (Figure 5G and H).
AC012213.3 Gene Silencing Blocked Breast Cancer Proliferation and Invasion Through the Downregulation of RAD54B and PI3K/AKT Signaling Activity
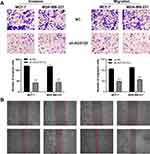
We hypothesized that AC012213.3 might be mediating cancer progression through the RAD54B protein. To understand the interaction between the two genes, we overexpressed RAD54B in the AC012213.3 knockdown cells. qRT-PCR analysis showed sufficient RAD54B overexpression in MCF-7 and MDA-MB-231 cells (Figure 6A and B). As expected the CCK8 and colony formation and transwell assays indicated that, whereas suppression of AC012213.3 could inhibit cell proliferation, overexpression of the RAD54B restores the proliferation phenotype (Figure 6C–F). PI3K/AKT signaling pathway has been shown to mediate development of many tumor cells. Here, we explored whether the AC012213.3 acts as an oncogene in breast cancer through the PI3K/AKT signaling pathway. The data showed that suppression of p-AKT and cyclinD1 proteins in the AC012213.3 knockdown cells (Figure 7A and B). However, overexpression of RAD54B in the AC012213.3 knockdown cells significantly increased the expression of p-AKT and cyclinD1. These findings indicated that the AC012213.3 could activate the PI3K/AKT pathway, and the activation might be partly dependent on the RAD54B expression.
Discussion
Breast cancer is the most common malignancy and the second cause of cancer-related deaths in women.2 With the development of medical technologies, there was increase in the five-year survival rate of breast cancer patients in the past decades.14 However, the treatment efficacy for advanced-stage breast cancer still remains unsatisfactory. Thus, there is an urgent need for more effective tumor biomarkers and therapeutic targets.
LncRNAs have been reported to play an essential role in tumor progression and development.15 For instance, lncRNA ZFPM2-AS1 has been shown to significantly accelerated hepatocellular carcinoma progression through a competitive endogenous RNA mechanism of miR-139/GDF10 axis (ceRNA).16 On the other hand, lncRNA LINC00460 promotes epithelial-mesenchymal transition (EMT) process by facilitating the movement of peroxiredoxin-1 into the nucleus,17 while lncRNA CYTOR has been shown to be an oncogene in promoting colon cancer metastasis.18 Our study identified a novel lncRNA AC012213.3 which is highly expressed in breast cancer tissues and cells and associated with patient survival. Our data showed that AC012213.3 might be an oncogene and presents a potential therapeutic target in breast cancer.
In addition, we showed that RAD54B was a downstream molecular of AC012213.3, and partly responsible for the cancer-promoting effect of AC012213.3. RAD54B is associated with homLologous recombination,19 and aberrant RAD54B expression might play a role in cancer. Since RAD54B ensures faithful and accurate repair of DNA damage, its overexpression and downregulation would perturb genomic instability which could confer oncogenic properties.20 Previous data has shown that suppression of RAD54B could block proliferation and promote apoptosis of liver and lung cancer cells.21,22 In addition, RAD54B could remarkably facilitate cancer progression in breast cancer.13
We showed that PI3K/AKT signaling pathway might mediate the progression effect by AC012213.3 and RAD54B. PI3K/AKT signaling participates in the pathological and physiological processes of cells and is regarded as a classical cancer-related pathway.23 Aberrant activation of AKT was observed in multiple cancers such as lung, ovarian and pancreatic cancer.24 A previous study showed that exosomal MALAT1 could promote the invasion and metastasis of colorectal cancer by enhancing fucosylation and PI3K/Akt pathway.25 In addition, another study showed that SIK2 could impact metabolic reprogramming in ovarian cancer through PI3K/AKT/HIF-1α pathway.26 Similarly, drugs targeting PI3K/AKT pathway have been developed and evaluated in preclinical studies.27
Here, using bioinformatics analysis, we identified AC012213.3 and showed that it is highly expressed in breast cancer tissues and cell lines. In addition, we showed that the suppression of AC012213.3 significantly inhibited cell proliferation and promoted cell apoptosis in MCF-7 and MDA-MB-231 cell lines. Thus, AC012213.3 might facilitate cell metastasis. Besides, RAD54B was also shown to be involved in the progression of breast cancer. Further assays showed that overexpression of RAD54B could partially reverse the tumor-inhibition effect caused by the suppression of AC012213.3. In addition, Western blot analysis demonstrated that AC012213.3 might promote breast cancer progression through PI3K/AKT pathway.
Conclusion
Taken together, our data showed that lncRNA AC012213.3 was highly expressed in the breast cancer tissues and was associated with patient survival. In addition, the oncogenic effect of AC012213.3 was partially dependent on the RAD54B/PI3K/AKT axis.
Funding
This study was supported by Chengdu High-level Key Clinical Specialty Construction Project; Project of Sichuan Medical Association (S17042); China International Medical Exchange Foundation (Z-2014-06-19389); Project of Beijing CSCO Clinical Oncology Research Foundation (Y-MSD2020-0406); Chengdu Municipal Health and Family Planning Commission (2018049); Scientific Research Project of Xinglin Scholars of Chengdu University of Traditional Chinese Medicine (YYZX2020021).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
2. Veronesi U, Boyle P, Goldhirsch A, Orecchia R, Viale G. Breast cancer. Lancet. 2005;365(9472):1727–1741. doi:10.1016/S0140-6736(05)66546-4
3. Olivotto IA, Bajdik CD, Ravdin PM, et al. Population-based validation of the prognostic model ADJUVANT! For early breast cancer. J Clin Oncol. 2005;23(12):2716–2725. doi:10.1200/JCO.2005.06.178
4. Cai P, Otten ABC, Cheng B, et al. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res. 2020;30(1):22–34. doi:10.1101/gr.251561.119
5. Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42(5):792–804. doi:10.1016/j.immuni.2015.05.004
6. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
7. Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. doi:10.1038/s41556-019-0299-0
8. Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234(6):9105–9117. doi:10.1002/jcp.27587
9. Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11(1):5. doi:10.1186/1476-4598-11-5
10. Xiu B, Chi Y, Liu L, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18(1):187. doi:10.1186/s12943-019-1115-y
11. Wang Y, Wu S, Zhu X, et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med. 2020;217(3):e20190950. doi:10.1084/jem.20190950
12. Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746–2755. doi:10.1038/onc.2015.340
13. Zhang Z, Li X, Han Y, et al. RAD54B potentiates tumor growth and predicts poor prognosis of patients with luminal A breast cancer. Biomed Pharmacother. 2019;118:109341. doi:10.1016/j.biopha.2019.109341
14. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448.
15. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
16. He H, Wang Y, Ye P, et al. Long noncoding RNA ZFPM2-AS1 acts as a miRNA sponge and promotes cell invasion through regulation of miR-139/GDF10 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):159. doi:10.1186/s13046-020-01664-1
17. Jiang Y, Cao W, Wu K, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365. doi:10.1186/s13046-019-1364-z
18. Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–1298. doi:10.1016/j.ymthe.2018.02.024
19. Yasuhara T, Suzuki T, Katsura M, Miyagawa K. Rad54B serves as a scaffold in the DNA damage response that limits checkpoint strength. Nat Commun. 2014;5:5426. doi:10.1038/ncomms6426
20. McAndrew EN, McManus KJ. The enigmatic oncogene and tumor suppressor-like properties of RAD54B: insights into genome instability and cancer. Genes Chromosomes Cancer. 2017;56(7):513–523. doi:10.1002/gcc.22458
21. Wang R, Li Y, Chen Y, et al. Inhibition of RAD54B suppresses proliferation and promotes apoptosis in hepatoma cells. Oncol Rep. 2018;40(3):1233–1242.
22. Xu C, Liu D, Mei H, Hu J, Luo M. Knockdown of RAD54B expression reduces cell proliferation and induces apoptosis in lung cancer cells. J Int Med Res. 2019;47(11):5650–5659. doi:10.1177/0300060519869423
23. Yang Q, Jiang W, Hou P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol. 2019;59:112–124. doi:10.1016/j.semcancer.2019.04.001
24. Song M, Bode AM, Dong Z, Lee MH. AKT as a therapeutic target for cancer. Cancer Res. 2019;79(6):1019–1031. doi:10.1158/0008-5472.CAN-18-2738
25. Xu J, Xiao Y, Liu B, et al. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer Res. 2020;39(1):54. doi:10.1186/s13046-020-01562-6
26. Gao T, Zhang X, Zhao J, et al. SIK2 promotes reprogramming of glucose metabolism through PI3K/AKT/HIF-1α pathway and Drp1-mediated mitochondrial fission in ovarian cancer. Cancer Lett. 2020;469:89–101. doi:10.1016/j.canlet.2019.10.029
27. Guerrero-Zotano A, Mayer IA, Arteaga CL. PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016;35(4):515–524. doi:10.1007/s10555-016-9637-x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.