Back to Journals » OncoTargets and Therapy » Volume 13
Overexpression of RNF126 Promotes the Development of Colorectal Cancer via Enhancing p53 Ubiquitination and Degradation
Authors Wang S , Wang T , Wang L , Zhong L, Li K
Received 12 July 2020
Accepted for publication 27 September 2020
Published 28 October 2020 Volume 2020:13 Pages 10917—10929
DOI https://doi.org/10.2147/OTT.S271855
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Shiyang Wang, 1, 2 Tianlong Wang, 1 Li Wang, 1 Liansheng Zhong, 3 Kai Li 2
1Department of Geriatric Surgery, The First Hospital, China Medical University, Shenyang 110001, People’s Republic of China; 2Department of Surgical Oncology, The First Hospital, China Medical University, Shenyang 110001, People’s Republic of China; 3Department of Bioinformatics, College of Life Science, China Medical University, Shenyang 110001, People’s Republic of China
Correspondence: Kai Li
Department of Surgical Oncology, The First Hospital, China Medical University, Shenyang 110001, People’s Republic of China
Tel +86-13998245233
Email [email protected]
Background: RING finger protein 126 (RNF126), as a novel E3 ubiquitin ligase, plays an oncogenic role in several solid cancers. But its potential role in colorectal cancer (CRC) that harbored 50% mutant p53, to our knowledge, is rarely reported.
Materials and Methods: We investigated the clinical significance and relationship of RNF126 and p53 in CRC tissues and cells. Meanwhile, WB, qRT-PCR, co-IP, MTT, and transwell were used to investigate the function and molecular mechanism of RNF126 in regulating malignant biology in vitro.
Results: RNF126 was overexpressed in human CRC specimens, which was tightly associated with tumor size (P=0.021), T stage (P=0.030), lymph node metastasis (P=0.006), TNM stage (P=0.001), and the poor survival (P=0.003) of CRC patients. RNF126 had no association with p53 mutation in CRC specimens, and in p53 mutant Colo-205 and SW620 cells. However, in p53 wildtype HCT116 and HCT-8 cells, RNF126 silencing upregulated p53 and p21 but inhibited Rb phosphorylation at Serine 780 (pRb), which was inhibited by p53siRNA. Conversely, RNF126 overexpression downregulated p53 and p21 but promoted pRb expression, which was reversed by a classic proteasome inhibitor, MG132. However, the mRNA levels of above target genes were unchanged, implying a ubiquitination dependent post-translational modification involving in above regulation. Meanwhile, RNF126 was co-immunoprecipitated with p53 and p21 to form a triple complex. RNF126 silencing and overexpression inhibited and promoted p53 ubiquitination and degradation in vitro, respectively. In addition, p53siRNA reversed RNF126 silencing-inhibited cell proliferation, drug resistance, and cell mobility in HCT116 cells. Conversely, MG132 inhibited RNF126 overexpression-promoted above cell biology in HCT-8 cells.
Conclusion: Overexpression of RNF126 was remarkably associated with multiple advanced clinical characters of CRC patients independent of mutant p53. RNF126 promotes cell proliferation, mobility, and drug resistance in CRC via enhancing p53 ubiquitination and degradation.
Keywords: RING finger protein 126, p53, ubiquitination, colorectal cancer
Corrigendum for this paper has been published
Introduction
Colorectal cancer (CRC) is one of the most common cancers over the world, with the fourth highest mortality rate, following lung, breast, and prostate cancer.1 In China, the morbidity and mortality of CRC ranked top five in both male and female population.2 The dysregulation of various molecular signal pathways involving the imbalance of oncogene and tumor suppressor has been attributed to the progression of CRC.3 However, the ubiquitin–proteasome system is one of the key post-translational modifications in regulating the tumor signaling pathway.4,5 As the second most prevalent posttranslational modification, ubiquitination is the covalent attachment of ubiquitin to target proteins via a cascade of enzymatic reactions, including Ub-activating (E1), Ub-conjugating (E2), and Ub-ligating (E3) enzymes. Accumulating evidence suggests that alterations in the activity of E3 ligases play a significant role in human malignancies.6
Ring finger protein 126 (RNF126) is identified as a novel E3 ubiquitin ligase, which contains a Ring finger and a Zn finger at the C- and N-terminus. It plays a crucial role in regulating cell cycle, proliferation, and DNA damage response.7–9 Recently, mounting studies show an oncogenic role of RNF126 in several cancers, including breast,10 gastric,11 prostate,7 and ovarian cancers.12 However, its potential role in colorectal cancer (CRC) and the corresponding molecular mechanism, to our knowledge, has not been reported.
p53 mutation occurs in 40–50% of sporadic CRC, is the second key genetic step in CRC, which promotes the transition of adenomas into adenocarcinoma.13 It is well known that ubiquitination plays a major part in p53 regulation.14,15 Although Mdm2 is identified as a classic E3 ubiquitin ligase for p53, there are still an increasing number of other E3 ligases target p53 independent of Mdm2.16 In the current study, we systematically investigate the potential role and corresponding mechanism involving the interaction between RNF126 and p53 in the progression of CRC, which offers a promising gene target for CRC intervention.
Materials and Methods
Tissues, Cell Lines, and Reagents
Our research was approved by the institutional review board from the First Hospital of China Medical University. Each patient enrolled in this study signed an informed consent form for samples which contained the research and collection involving human biological samples or personal data. In total, 147 formalin-fixed and paraffin-embedded CRC and corresponding normal tissues were obtained from CRC patients at our own hospital between 2013 and 2018. The histologically adjacent normal tissues were only extracted from the mucosa that was at least 5 cm away from the cancer. An additional 24 CRC fresh tissues were collected for late Western blot (WB) and Real-time quantitative PCR (qRT-PCR) assays.
Human HCT116, HCT-8, Colo205, and SW620 cell lines were purchased from the cell bank of Chinese academic sciences (Shanghai, China). These cell lines were maintained in recommended growth media with 10% fetal calf serum (GIBCO, Carlsbad, CA, USA) and were authenticated for contamination prior to the experiment in vitro.
Immunohistochemistry Assays
IHC was conducted according to the previous study.17 The formalin fixed CRC specimens were made into a paraffin block and 4-µm sections. Then the slices were deparaffinized and dehydrated, covered with peroxyacetic acid, sent to antigen retrieval, blocked with 10% goat serum, and incubated with anti-RNF126 (Abcam, Cambridge, UK, dilution: 1:100) and p53 (Proteintech, Chicago, IL, USA, dilution: 1:100). All sections were washed with PBS three times and incubated with the streptavidin–HRP tagged secondary antibody, detected with DAB, co-stained with haematoxylin, and finally evaluated by two professional pathologists. Staining intensity was recorded as negative to strong (0–3), while staining area was scored as 0–4 (<5%; 10–25%; 26–50%; 51–75%; >76%). The scores were summed together as the final staining scores (0–7). Tumors having a final staining score ≥3.5 were considered to be RNF126 and p53 high expression.
Western Blot
The total proteins extracted from the lysates of CRC cells and tissues were implanted into 12% SDS-polyacrylamide gels, transferred to PVDF membrane, blocked with 5% BSA, and incubated with primary antibodies: RNF126 (Abcam, dilution: 1:1,000), p53 (Proteintech, dilution: 1:1,000), p21 (Cell Signaling Technology, Beverly, USA, dilution: 1:500), Retinoblastoma protein (Rb, Abcam, dilution: 1:1,000), pRb (phosphor- S780, Abcam, dilution: 1:500), and GAPDH (Proteintech, dilution: 1:3,000). Then all the bands were covered with HRP tagged secondary antibodies (Proteintech) and were detected with the ECL machine (Bio-Rad, CA, USA). Cells were pretreated with 2 µM of proteasome inhibitor MG132 (Abcam, ab141003) for 18 hours prior to WB under corresponding standard protocol. WB was repeated at least three times.
Total RNA Extraction and Real-Time Quantitative PCR (qRT-PCR)
Total RNA was extracted from CRC tissue specimens and four CRC cell lines with TRIZOL reagent, as described by the manufacturer. The A260/A280 ratios of all RNA samples ranged from 1.8t to 2.0. cDNA was synthesized from approximately 500–1,000 ng of total RNA using the High Capacity cDNA Reverse Transcriptase Kit (Takara Bio, Japan). The cDNA synthesis reaction was performed using a MyCycler Thermal Cycler (Bio-Rad) using the following conditions: 25°C, 10 minutes; 37°C, 120 minutes; 85°C, 5 minutes; 4°C, hold. cDNA samples were diluted 1:4 using nuclease-free water. Two microliters of cDNA were used as a template for qPCR. After reaction assembly, the plate was centrifuged briefly. Samples were performed in duplicate. Reaction mixtures were prepared as follows: 94°C for 40 seconds and 45 cycles of 94°C for 5 seconds and 58°C for 30 seconds, and were analyzed in a Light Cycler 2.0 with SYBR Premix Ex TaqTM II kit (Takara Bio). The primer sequences: RNF126, 5ʹ- CCCACGCTGGAAGGGATCAT G‐3ʹ and 5ʹ- CCTACGTGCTCCTCAGTGAC-3ʹ; p53, 5ʹ- AGAGCTGAATGAGGCCTTGGAA-3ʹ and 5ʹ- GAGTCAGGCCCTTCTGTCTTGAAC-3ʹ; p21, 5ʹ- AGGTGGACCTGGAGACTCTCAG-3ʹ and 5ʹ-TCCTCTTGGAGAAGATCAGCCG-3ʹ. GADPH, 5ʹ- TGACTTCAACAGCGACACCCA −3ʹ and 5ʹ - CACCCTGTTGCTGTAGCCAAA −3ʹ. Quality control of the PCR products was monitored with melt-curve analysis. The results were calculated using the -ΔΔCt method.
Co-immunoprecipitation
For co-immunoprecipitation (Co-IP), the whole protein lysates prepared from HCT116 and HCT-8 cells were extracted in a RIPA lysis buffer. Briefly, RNF126 and p53 antibodies were first mixed with magnetic-beads (Thermo scientific, Rockford, IL, USA) for at least 4 hours. Antibody-beads complexes were then mixed with the supernatants of protein lysates with a rotator overnight. The final antibody–protein immunocomplex was stripped by boiling with 5x loading sample buffer for WB incubated with RNF126, p53, p21, and GAPDH antibodies. Input and IgG panels were used as the positive and negative control, respectively. Co-IP was repeated at least three times. For ubiquitination assays, we routinely took p53 as the Co-IP target (antibody-protein immunocomplex) that was incubated with anti-Ubiquitin antibody (Abcam) for late Ubiquitin detecting.
RNA Interference and Plasmid
Two RNF126siRNA (si1-RNF126 and si2-RNF126) sequences were : 1) GAGTCCTGCACTCAAACCCTATGGA; 2) CACTCAAACCCTATGGACTACGCCT. According to a previous study,18 the siRNA sequence for p53 was: GACUCCAGUGGUAAUCUACTT. The pcDNA3-EGFP plasmid (RNF-126-GFP) and corresponding empty plasmid (GFP) were synthesized from Genechem (Shanghai, China). siRNA and plasmids were mixed with Oligofectamine 3000 (Invitrogen, Carlsbad, CA, USA) for transfection under standard protocol. HCT116, Colo-205, and SW620 with RNF126 high expression were used for RNF126 silencing, while HCT-8 cells with low RNF126 were used for RNF126 overexpression.
MTT Assays
MTT assay was first conducted to analyze the role of RNF126 in cell proliferation at different time points (1–5 days) combining with or without p53siRNA or MG132. Briefly, si1-RNF126 or siRNA control (siCtrl) transfected HCT116 cells were first co-transected with p53siRNA for 24 hours and were then harvested with 0.05% trypsin/EDTA, seeded into 96-well plates at the density of 5,000 viable cells per well and finally incubated at 1–5 days. At the end of each time point, 10 µL of MTT (5 mg/mL in PBS, Sigma) was added and the cells were incubated for 4 hours at 37°C. Then 100 µL of dimethyl sulfoxide (Sigma) was added to each well for 20 minutes per experiment group at a wavelength of 570 nm in an ELISA 96-well microtiter plate reader (Bio-rad, USA). Similarly, RNF126-GFP and GFP transfected HCT-8 cells were pre-treated with 2 µM of MG132 18 hours prior to MTT assays. For drug resistance assays, the above cell lines were cultured into 96-well plates overnight and then added with 5-Fluorouracil (5-Fu, Sigma-Aldrich) and Oxaliplatin (OXA, Abcam) twice within 2 days at various concentrations shown in the result sections. Experiments were performed in triplicate.
Cell Mobility Assays
si1-RNF126/siCtrl transfected HCT116 cells (with or without p53siRNA) and RNF126-GFP/GFP transfected HCT-8 cells (with or without 2 µM of MG132) were implanted into 12.0-µM pore size membrane inserts (Corning Inc., NY, USA) coated with matrigel (Corning Inc) in 24-well plates with free serum media. Media plus 10% FBS was put into the bottom as a stimulus. Twenty-four hours later, the migrated cells from upside to the underside of the inserts were fixed and co-stained with crystal violet (Sigma). The final cell migrated number was calculated in five random fields per each well under a microscope (20x magnification). The migration assay was implemented in a similar way without matrigel. Cell motility assays were repeated at least three times.
Statistical Analysis
Statistical analysis was performed using SPSS software 20.0 (SPSS Inc., Chicago, IL, USA). For IHC assays, the difference of RNF126 expression in CRC and paired normal tissues and the corresponding clinicopathological significance with CRC patients were analyzed via paired non-parametric test and chi-squared test, respectively. The Kaplan-Meier method and the Log rank test was used to estimate the survival data. RNF126 differential expression in CRC and paired normal tissues by WB and qRT-PCR was analyzed via paired sample t-test. All data in vitro were expressed as means±SE and were compared through independent sample t-test. P<0.05 was considered to be statistically significant.
Results
The Clinical Significance of RNF126 and p53 Expression in CRC Specimens
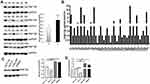
RNF126 was mainly expressed in the cytoplasm in both CRC and paired adjacent colorectal tissues (Figure 1A and B), while p53 was expressed in nuclear CRC specimens (Figure 1C and D). RNF126 was overexpressed in CRC specimens compared with paired adjacent normal tissues (54.4%, 80/147 vs 30.6%, 45/147, P<0.01) (Figure 1A and B). P53 detected by IHC was regarded as p53 mutation because of the long half-life time in p53 mutant status.19 Overexpression of p53 in CRC specimens (51%, 75/147) had no correlation with RNF126 by correlation analysis (r=−0.114, P=0.166) (Table 1). Just as shown in Figure 1C and D, most serial CRC specimens with p53 overexpression followed with both RNF126 high- and low-staining.
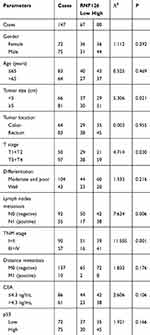 |
Table 1 The Relationship of RNF126 Overexpression with Clinical Data |
The relationship of RNF126 overexpression with clinical data is summarized in Table 1. RNF126 overexpression was tightly associated with tumor size (P=0.021), T stage (P=0.030), lymph node metastasis (P=0.006), and TNM stage (P=0.001) of CRC patients, but had no association with other parameters, including gender, age, tumor location, differentiation, distant metastasis, and CEA. In Kaplan-Meier assay, CRC patients with high RNF126 expression had a worse overall survival (P=0.003) (Figure 1E). p53 overexpression was also associated with poor survival (Figure 1F), which was consistent with a previous study.20
In addition, WB and qRT-PCR further verified that RNF126 protein and mRNA levels were significantly higher in 24 CRC tissues than in the paired normal colorectum (P<0.01 and P<0.01, respectively) (Figure 2A and B).
Taken together, overexpression of RNF126 was associated with multiple clinical characters in CRC patients independent of mtp53, which drove us to investigate the potential interaction of RNF126 with p53 in vitro.
The Relationship of RNF126 and p53 in vitro
Four CRC cell lines with different p53 status were used in vitro. HCT116 and HCT-8 CRC cell lines contained wildtype p53 (wtp53),21 while Colo-205 and SW620 cells harbored mutant p53 (mtp53).22
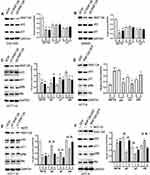
RNF126 protein and mRNA expression was higher in HCT116, Colo-205, and SW620 cells compared with HCT-8 cells. Thus, HCT116, Colo-205, and SW620 with high-RNF126 expression were used for silencing RNF126, while HCT-8 cells with low-RNF126 were used for overexpressing RNF126 (Figure 2C and D).
We first showed that RNF126 expression was significantly decreased in si1-RNF126 and si2-RNF126 groups compared with the siCtrl group in mtp53.
Colo-205 and SW620 cells, and wtp53 HCT116 (Figure 3A–C), whereas RNF126 expression was much higher in the RNF126-GFP group in contrast with the GFP group in wtp53 HCT-8 cells (Figure 3D). RNF126 silencing did not change p53 and its downstream target p21 expression in mtp53 Colo-205 and SW620 cells (Figure 3A and B). However, p53 and p21 expression was significantly increased, but phosphorylated Retinoblastoma protein (pRb, the downstream of p53/p21) was decreased in RNF126 silencing HCT116 cells (Figure 3C). Total Rb protein level was unchanged in each group. Moreover, p53siRNA reversed RNF126 silencing-promoted p53 and p21, and RNF126 silencing-inhibited pRb expression (Figure 3E), which indicated a close interaction between RNF126 and p53-p21-Rb signaling. As mentioned above, Mdm2 independent ubiquitination also plays a significant role in mediating p53 regulation. As a novel E3 ligase, p53 and p21 expression was significantly decreased, but pRb was increased in RNF126-GFP transfected HCT-8 cells (Figure 3D), which was reversed by as a classic proteasome inhibitor, MG132 (Figure 3F). Meanwhile, MG132 also increased the total Rb protein level in the same cell line (Figure 3F). However, both RNF126 silencing and overexpression had no effect in the mRNA levels of p53 and p21 (Supplemental Figure 1A and B). Similarly, MG-132 had no effect in the mRNA levels of RNF126, p21, and p53 in vitro (Supplemental Figure 1C), which further verified that MG-132 mediated the interaction between RNF126 and p53/p21 via the ubiquitination dependent posttranslational modification.
Taken together, a potential interaction between RNF126 and wtp53-p21-Rb signaling via the ubiquitination system in vitro has driven us to take in-depth analyses of the molecular mechanism next.
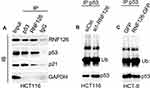
RNF126 Promoted P53 Ubiquitination and Degradation
We further showed that RNF126 was co-immunoprecipitated with p53 and p21 in high-RNF126 expression HCT116 cells (Figure 4A). Similarly, p53 was also co-immunoprecipitated with RNF126 and p21 when using p53-lysates immunocomplex as the immunoprecipitated target. The triple complex was formed in vitro (Figure 4A). Moreover, we detected the weakened ubiquitination of p53 from the p53-lysates immunocomplex in si1-RNF126 transfected HCT116 cells compared with siCtrl groups following the increase of p53 protein expression (Figure 4B). Conversely, the enhanced ubiquitination of p53 was observed in the p53-lysates immunocomplex in RNF126-GFP transfected HCT-8 cells compared with GFP groups following the decrease of p53 protein level (Figure 4C). Taken together, RNF126 promoted p53 ubiquitination and degradation, which drove us to further investigate their coordinate function in malignant biology in vitro.
RNF126 Regulated Cell Proliferation and Drug Resistance in vitro in p53 Dependent-Ubiquitination
p53 was a key regulator in cell cycle, proliferation, apoptosis, and chemo- and radio-sensitivity of several cancers.23 We inferred RNF126 regulated cell proliferation and drug resistance in p53 dependent-ubiquitination.
In MTT assays, RNF126 silencing inhibited cell proliferation in HCT116 cells in a time dependent manner. Cell growth in the si1-RNF126 group was significantly decreased compared with the siCtrl group at the culturing time points of 4 to 5 days (Figure 5A). However, p53siRNA restored RNF126 silencing-inhibited cell proliferation at the same time points (Figure 5A). Conversely, RNF126 overexpression promoted cell proliferation in HCT-8 cells when constantly observed cell growth at 4 to 5 days, which was also significantly inhibited by 2 µM of MG132 (Figure 5B).
5-Fu and OXA are identified as the first-line chemotherapy for metastatic or recurrent gastrointestinal cancer patients.24 Because RNF126 silencing or overexpression alone had no effect in cell proliferation less than 3 days based on the above results, we treated CRC cells with various concentration of GEM and OXA only for 2 days to exclude its own effect in cell validity. In HCT116 cells, cell proliferation in the si1-RNF126 group was significantly decreased in contrast with the siCtrl group under various concentrations of 5-Fu (12.5–50 µg/mL) treatment, which was reversed by p53siRNA (Figure 5C). The similar results were also observed in HCT116 cells under various concentration (25–100 µg/mL) of OXA treatment (Figure 5D). Conversely, cell proliferation in RNF126-GFP group of HCT-8 cells was significantly increased in contrast with the GFP group with various concentrations of 5-Fu (5–20 µg/mL) treatment. However, MG132 reversed RNF126 overexpression-promoted 5-Fu resistance at the same concentration point (Figure 5E). The similar results were also observed in HCT-8 cells under OXA treatment (20–40 µg/mL) (Figure 5F).
Taken together, RNF126 improved cell proliferation and drug resistance in vitro in p53 dependent-ubiquitination.
RNF126 Promoted Cell Mobility in vitro in p53 Dependent-Ubiquitination
Migration and invasion are prerequisites for CRC progression and metastasis, which is prevalently regulated by p53.25 In the present study, cell invasion (Figure 6A) and migration (Figure 6B) in si1-RNF126 transfected HCT116 cells were significantly decreased in contrast with the siCtrl group, which was significantly reversed by p53siRNA (Figure 6A and B). Conversely, RNF126 promoted cell invasion (Figure 6C) and migration (Figure 6D) in HCT-8 cells compared with the GFP group. However, MG132 significantly inhibited RNF126-induced cell mobility in vitro (Figure 6C and D). Based on the above results, RNF126 inhibited cell mobility in vitro in p53 dependent-ubiquitination.
Discussion
Regarding any dysregulation of ubiquitination is tightly linked to cancer development,26 we investigate a novel E3 ligase RNF126 and its potential ubiquitin targets in CRC development. For the first time, we found that overexpression of RNF126 was tightly associated with multiple clinical characters of CRC patients independent of mtp53. RNF126 regulated wtp53 and its downstream target p21 and Rb via activating the ubiquitin-proteasome system, which coordinately mediated cell malignant biology of CRC cells.
We first found overexpression of RNF126 in human CRC specimens was tightly associated with tumor size, T stage, lymph node metastasis, TNM stage, and the poor survival of CRC patients. The clinical significance of RNF126 expression was also reported in other solid cancers. RNF126 overexpression was remarkably associated with lymph node metastasis, pathological differentiation, and FIGO stage in ovarian cancer,12 and was associated with a poor clinical outcome in breast and gastric cancers.10,11 P53 is well known as a key hallmark in CRC with approximately 40–50% mutation.13 Though mtp53 was associated with poor prognosis of CRC patients, no remarkable relationship between RNF126 and mtp53 was found in the current study. p53 is a key target by ubiquitin as well as ubiquitin-like proteins. Ubiquitin dependent modifications are critical for the function of p53 involving the control of its degradation as well as localization and activity.27 RNF126, as one of the RING family of ligases, contains a zinc-binding RING finger domain that performs the ubiquitin ligase activity. Thus, we reasoned that RNF126 regulated the ubiquitination of wtp53 in vitro.
Consistent with the results in tissue level, RNF126 silencing had no effect in mtp53 in Colo-205 and SW620 cells. However, p53 silencing reversed RNF126 silencing-induced p53 and p21, and RNF126 silencing-inhibited pRb protein expression in wtp53 HCT116 cells, implying a specific regulation of RNF126 toward p53-p21-Rb signaling. Moreover, MG132 reversed RNF126 overexpression-inhibited p53 and p21, and RNF126 overexpression-promoted pRb expression in wtp53 HCT-8 cells. However, the mRNA levels of above target genes were unchanged in vitro, which further verified a ubiquitin–proteasome system-dependent post-translational modification involving the above regulation. Further molecular mechanism first showed that RNF126, p53, and p21 formed a triple complex. RNF126 silencing or overexpression weakened or promoted p53 ubiquitination following with the increase and decrease of p53 protein expression, respectively. Taken together, RNF126 promoted p53 ubiquitination and degradation, which, to our knowledge, has not been reported previously. RNF126 also targets a variety of other proteins for ubiquitin-dependent degradation. For example, RNF126 promotes completion of DNA repair by ubiquitylating Ku80 and releasing Ku70/80 from damaged DNA.28 RNF126 contributes to BAG6-mediated quality control,8 and has been shown to ubiquitinate activation-induced cytidine deaminase.29 RNF126 also targets p21 for ubiquitin-mediated degradation in breast and prostate cancer cells.7 Taken together, RNF126 is identified as a new E3 ligase target p53 independent of Mdm2, which has not been reported previously.
p21 is a key factor mediated by p53 in response to DNA damage30 and cell cycle transition (G1 to S phase).31 Meanwhile, the p53/p21 complex rather than p53 itself regulates cell invasion and death by targeting Bcl-2 proteins.32 p53 and p21 cooperate to promote the Mdm2-dependent degradation of Slug, in turn suppressing lung cancer cell invasion.33 Similarly,
Rb, acts as the potential downstream of p53 or p21 signal pathway,34–36 and plays a significant role in cell invasion and proliferation of several cancer cells.37–39 p21 induction leads to Rb dephosphorylation and activation, with ensuing G1 arrest. Inactivation or loss of Rb in stromal fibroblasts promotes epithelial cell invasion37 and prostate cancer metastasis.38 Dephosphorylation of the Rb, which leads to the activation of Rb, inhibits cancer cell EMT via Zeb.39 The Rb also plays a pivotal role in the negative control of the cell cycle and in tumor progression.40 Regarding the key role of p53-p21-Rb signaling in cell proliferation and mobility, we finally found that RNF126 promoted cell proliferation, drug resistance, and cell mobility in vitro via inhibiting p53/p21 in ubiquitin-dependent degradation.
RNF126 also promotes cell proliferation by enhancing p21 ubiquitination in MDA-MB-231 and PC3 cancer cells,7 whereas RNF126 silencing inhibits the proliferation and viability of SCC9 and SCC25 cells via decreasing the activity of AKT1.41 For the first time, we found a coordinate regulation between RNF126 and p53-p21-Rb signaling in mediating cell biology in CRC via ubiquitin-dependent degradation.
In conclusion, overexpression of RNF126 was remarkably associated with multiple advanced clinical characters of CRC patients independent of mtp53. RNF126 negatively regulated wtp53 and its downstream target p21 and Rb via activating the ubiquitin-proteasome system. The close interaction between RNF126 and p53-p21-Rb signaling coordinately promoted cell proliferation, mobility, and drug resistance in vitro. The small molecular inhibitor of RNF126 will be designed and used in vivo in our future study, which would be a promising gene target for CRC intervention.
Disclosure
The authors declare no conflict of interest.
References
1. Global Cancer Observatory. Available from: http://gco.iarc.fr/.
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China. CA Cancer J Clin. 2015;66(2):115–132.
3. Koveitypour Z, Panahi F, Vakilian M. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019;9:97. doi:10.1186/s13578-019-0361-4
4. Zeng K, Chen X, Hu X, et al. LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2-mediated p53 ubiquitination and degradation. Oncogene. 2018;37(41):5534–5551. doi:10.1038/s41388-018-0352-7
5. Huang C, Wu S, Li W, et al. Zinc-finger protein p52-ZER6 accelerates colorectal cancer cell proliferation and tumour progression through promoting p53 ubiquitination. EBioMedicine. 2019;48:248–263. doi:10.1016/j.ebiom.2019.08.070
6. Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi:10.1038/nrc1994
7. Zhi X, Zhao D, Wang Z, et al. E3 ubiquitin ligase RNF126 promotes cancer cell proliferation by targeting the tumor suppressor p21 for ubiquitin-mediated degradation. Cancer Res. 2013;73(1):385–394. doi:10.1158/0008-5472.CAN-12-0562
8. Rodrigo-Brenni MC, Gutierrez E, Hegde RS. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell. 2014;55(2):227–237. doi:10.1016/j.molcel.2014.05.025
9. Zhang L, Wang Z, Shi R, et al. RNF126 quenches RNF168 function in the DNA damage response. Genomics Proteomics Bioinformatics. 2018;16(6):428–438. doi:10.1016/j.gpb.2018.07.004
10. Yang X, Pan Y, Qiu Z, et al. RNF126 as a biomarker of a poor prognosis in invasive breast cancer and CHEK1 inhibitor efficacy in breast cancer cells. Clin Cancer Res. 2018;24(7):1629–1643. doi:10.1158/1078-0432.CCR-17-2242
11. Migita K, Matsumoto S, Wakatsuki K, et al. RNF126 as a marker of prognosis and proliferation of gastric cancer. Anticancer Res. 2020;40(3):1367–1374. doi:10.21873/anticanres.14078
12. Wang C, Wen A, Qiao J, Liu Y, Guo Y, Wang W. High expression of RING finger protein 126 predicts unfavorable prognosis of epithelial ovarian cancer. Med Sci Monit. 2020;26:e921370.
13. Li H, Zhang J, Tong JHM, et al. Targeting the oncogenic p53 mutants in colorectal cancer and other solid tumors. Int J Mol Sci. 2019;20(23):5999. doi:10.3390/ijms20235999
14. Sakamoto KM. Ubiquitin-dependent proteolysis: its role in human diseases and the design of therapeutic strategies. Mol Genet Metab. 2002;77(1–2):44–56. doi:10.1016/S1096-7192(02)00146-4
15. Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17(1):86–92. doi:10.1038/cdd.2009.77
16. Coutts AS, Adams CJ, La Thangue NB. p53 ubiquitination by Mdm2: a never ending tail? DNA Repair (Amst). 2009;8(4):483–490. doi:10.1016/j.dnarep.2009.01.008
17. Huang L, Chen S, Fan H, Ai F, Sheng W. BZW2 promotes the malignant progression of colorectal cancer via activating the ERK/MAPK pathway. J Cell Physiol. 2020;235(5):4834–4842. doi:10.1002/jcp.29361
18. Sheng W, Dong M, Chen C, Li Y, Liu Q, Dong Q. Musashi2 promotes the development and progression of pancreatic cancer by down-regulating Numb protein. Oncotarget. 2017;8(9):14359–14373. doi:10.18632/oncotarget.8736
19. Kaserer K, Schmaus J, Bethge U, et al. Staining patterns of p53 immunohistochemistry and their biological significance in colorectal cancer. J Pathol. 2000;190(4):450–456. doi:10.1002/(SICI)1096-9896(200003)190:4<450::AID-PATH545>3.0.CO;2-8
20. Hamelin R, Laurent-Puig P, Olschwang S, et al. Association of p53 mutations with short survival in colorectal cancer. Gastroenterology. 1994;106(1):42–48. doi:10.1016/S0016-5085(94)94217-X
21. Vilgelm AE, Washington MK, Wei J, Chen H, Prassolov VS, Zaika AI. Interactions of the p53 protein family in cellular stress response in gastrointestinal tumors. Mol Cancer Ther. 2010;9(3):693–705. doi:10.1158/1535-7163.MCT-09-0912
22. Mariadason JM, Arango D, Shi Q, et al. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63(24):8791–8812.
23. El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22(47):7486–7495. doi:10.1038/sj.onc.1206949
24. Peinert S, Grothe W, Stein A, et al. Safety and efficacy of weekly 5-fluorouracil/folinic acid/oxaliplatin/irinotecan in the first-line treatment of gastrointestinal cancer. Ther Adv Med Oncol. 2010;2(3):161–174. doi:10.1177/1758834010365061
25. Sui X, Zhu J, Tang H, et al. p53 controls colorectal cancer cell invasion by inhibiting the NF-κB-mediated activation of Fascin. Oncotarget. 2015;6(26):22869–22879. doi:10.18632/oncotarget.5137
26. Voutsadakis IA. The ubiquitin-proteasome system in colorectal cancer. Biochim Biophys Acta. 2008;1782(12):800–808. doi:10.1016/j.bbadis.2008.06.007
27. Brooks CL, Gu W. p53 regulation by ubiquitin. FEBS Lett. 2011;585(18):2803–2809. doi:10.1016/j.febslet.2011.05.022
28. Shida N, Nakagawa T, Iemura SI, et al. Ubiquitylation of Ku80 by RNF126 promotes completion of nonhomologous end joining-mediated DNA repair. Mol Cell Biol. 2017;37(4):e00347–16.
29. Delker RK, Zhou Y, Strikoudis A, Stebbins CE, Papavasiliou FN. Solubility-based genetic screen identifies RING finger protein 126 as an E3 ligase for activation-induced cytidine deaminase. Proc Natl Acad Sci USA. 2013;110:1029–1034. doi:10.1073/pnas.1214538110
30. Hill R, Bodzak E, Blough MD, Lee PW. p53 binding to the p21 promoter is dependent on the nature of DNA damage. Cell Cycle. 2008;7(16):2535–2543. doi:10.4161/cc.7.16.6440
31. Pitolli C, Wang Y, Candi E, Shi Y, Melino G, Amelio I. p53-mediated tumor suppression: DNA-damage response and alternative mechanisms. Cancers. 2019;11(12):1983. doi:10.3390/cancers11121983
32. Kim EM, Jung CH, Kim J, Hwang SG, Park JK, Um HD. The p53/p21 complex regulates cancer cell invasion and apoptosis by targeting Bcl-2 family proteins. Cancer Res. 2017;77(11):3092–3100. doi:10.1158/0008-5472.CAN-16-2098
33. Kim J, Bae S, An S, et al. Cooperative actions of p21WAF1 and p53 induce slug protein degradation and suppress cell invasion. EMBO Rep. 2014;15(10):1062–1068. doi:10.15252/embr.201438587
34. Kramer DL, Vujcic S, Diegelman P, et al. Polyamine analogue induction of the p53-p21WAF1/CIP1-Rb pathway and G1 arrest in human melanoma cells. Cancer Res. 1999;59(6):1278–1286.
35. Benson EK, Mungamuri SK, Attie O, et al. p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene. 2014;33(30):3959–3969. doi:10.1038/onc.2013.378
36. Brugarolas J, Moberg K, Boyd SD, Taya Y, Jacks T, Lees JA. Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after gamma-irradiation. Proc Natl Acad Sci U S A. 1999;96(3):1002–1007. doi:10.1073/pnas.96.3.1002
37. Pickard A, Cichon AC, Barry A, et al. Inactivation of Rb in stromal fibroblasts promotes epithelial cell invasion. EMBO J. 2012;31(14):3092–3103. doi:10.1038/emboj.2012.153
38. Thangavel C, Boopathi E, Liu Y, et al. RB loss promotes prostate cancer metastasis. Cancer Res. 2017;77(4):982–995. doi:10.1158/0008-5472.CAN-16-1589
39. Egger JV, Lane MV, Antonucci LA, Dedi B, Krucher NA. Dephosphorylation of the Retinoblastoma protein (Rb) inhibits cancer cell EMT via Zeb. Cancer Biol Ther. 2016;17(11):1197–1205. doi:10.1080/15384047.2016.1235668
40. Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25(38):5220–5227. doi:10.1038/sj.onc.1209615
41. Wang L, Wang X, Zhao Y, et al. E3 ubiquitin ligase RNF126 regulates the progression of tongue cancer. Cancer Med. 2016;5(8):2043–2047. doi:10.1002/cam4.771
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.