Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
Overexpression of miR125b Promotes Osteoporosis Through miR-125b-TRAF6 Pathway in Postmenopausal Ovariectomized Rats
Authors Wang G, Zhang L, Yan C, Wang F, Zhang Y
Received 23 October 2020
Accepted for publication 19 December 2020
Published 15 February 2021 Volume 2021:14 Pages 671—682
DOI https://doi.org/10.2147/DMSO.S288338
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Konstantinos Tziomalos
Gang Wang, Lecheng Zhang, Chao Yan, Fengbin Wang, Yuelei Zhang
Department of Orthopedics, The First Affiliated Hospital of Anhui Medical University, Hefei, 230000, People’s Republic of China
Correspondence: Yuelei Zhang
Department of Orthopedics, The First Affiliated Hospital of Anhui Medical University, 218 Jixi Road, Hefei, 230000, People’s Republic of China
Tel/Fax +86-551-62922065
Email [email protected]
Background: Postmenopausal osteoporosis is one of the most common types of osteoporosis that women suffer from. Studies involving molecular mechanisms for designing better therapeutic strategies for postmenopausal osteoporosis are still rare. The present study investigates the role of miR-125b in postmenopausal osteoporosis.
Methods: Microarray analysis was done to screen the gene database. Tissue samples of postmenopausal women were collected to study the miRNA profiles. MC3T3-E1 cells were used and were submitted for transfection. CCK-8 assay was done to check the viability of cells, whereas toxicity was done by lactate dehydrogenase assay kit. TargetScan was done to target genes of miR-125b followed by confirmation by Luciferase reporter assay. For animal studies a rat model of ovariectomized rats was created. Bone mineral density and biomechanics were measured by densitometer. The mRNA levels were assessed by qRT-PCR and proteins by Western blot assay.
Results: miR-125b was over-expressed in human osteoporosis samples. In vitro studies suggested that miR-125b suppressed the cell viability and promoted release of LDH, it also enhanced the RANKL/OPG ratio and suppressed levels of BMP2 and Runx2. Bioinformatics identified TRAF6 as a potential target of miR-125b, further confirmed by luciferase assay, also miR-125b negatively regulated the levels of TRAF6 gene in osteoporosis bones involving the JAK2/STAT3 cascade. In the rat model, miR-125b decreased the bone mineral density and biomechanical parameters in bones by altering the TRAF6 gene involving the JAK2/STAT3 pathway.
Conclusion: The outcomes suggested that miR-125b was responsible for the development of postmenopausal osteoporosis and promoted its progression by the TRAF6 gene via the JAK2/STAT3 pathway.
Keywords: miR‐125b, osteoporosis, TRAF6, JAK2/STAT3 pathway
Introduction
Osteoporosis is a bone disorder featuring decreasing bone mineral density (BMD) and deterioration of the micro-architecture of bones contributing to the increased risk of fractures.1 It is already established that bones undergo the process of remodeling which includes formation of bone via generation of osteoblastogenesis and bone resorption by osteoclastogenesis.2 A very critical balance is needed between the process of bone formation and bone resorption for healthy bone mass, osteoporosis is just the imbalance between these two processes.3 Osteoporosis is of mainly two types, primary and secondary osteoporosis. The senile osteoporosis and postmenopausal osteoporosis are the primary types of osteoporosis, among them later one is the commonest type in women. Postmenopausal osteoporosis results from alterations in levels of estrogens, which leads to increased turnover of bones and finally leads to a loss of connectivity and porosity in the trabecular and cortical bones respectively.4 Recently, the number of subjects diagnosed with osteoporosis is double forwomen, compared to men.5 Hence a thorough study involving molecular mechanisms and the required therapeutic strategies for postmenopausal osteoporosis is needed.
MicroRNAs (miRs) are single stranded noncoding RNAs containing about 20–22 nucleotides. miRs are identified as potential regulators of various cellular processes such as cell proliferation, apoptosis and cell differentiation by targeting the 3ʹUTR segment of the target genes and alerting their expression.6 Recent studies have outlined thousands of miRs and multiple human genes involved in regulating them.7 A number of studies have emerged confirming the roles of various miRs responsible for regulating osteoporosis, miRs such as miR-542-3p were found to inhibit osteoporosis induced by ovariectomy in animal models involving SFRP18 miR-543 on the other hand was found to promote osteoporosis via the YAF-2 protein.9 Studies suggest the important role of miRs in the pathogenesis of osteoporosis and hence can be important therapeutic targets.10 Recently, miR-125b has been reported to be down regulated in the skin of systemic sclerosis resulting in apoptosis of dermal fibroblasts.11 miR-125b exerts a protective effect on heart tissue against acute myocardial infarction via p53 mediated apoptosis signaling involving TRAF6.12 A study recently reported that miR-125b inhibits the production of IL-6 in CD40-mediated B lymphocyte effector functions.13 However the regulatory role of miR-125b and the involved mechanism of miR-125b in osteoporosis remain unexplored.
In the present study, we screened the expression level of miR125b in osteoporosis tissue samples and also evaluated the effect of miR-125b on the process of bone resorption and formation. We also elucidated the mechanism underlying for miR-125b along with the target genes and the pathway involved in the development and progression of osteoporosis. For the in vitro studies we selected MC3T3-E1 cells and for in vivo analysis we developed an ovariectomized (OVX) rat model.
Materials and Methods
miRNA Expression Profile (Omnibus Database)
We screened the Omnibus database from the GDS928 dataset involving GSE1367 series, GDS1287 dataset involving GSE6711 series and the GDS3353 dataset with the GSE7429 series. We also extracted the miR-125b levels from the gene expression omnibus data. The GDS928 dataset platform with GSE1367 dataset was based on the Amersham Biosciences CodeLinkUniSet Mouse I Bioarray involving the screening of 1000 genes. The GDS1287 dataset with GSE6711 series was associated with Affymetrix Human Genome U133A Array involving 19 women. The GDS3353 dataset platform involving the GSE7429 series was based on Affymetrix Human Genome U133A Array which involved 20 women.
Clinical Blood Samples
The blood samples were collected from 18 postmenopausal women patients reported for osteoporosis visiting the Out-Patient Department (OPD) of the hospital between the periods February 2019 to April 2019. All 18 women submitted to physical examination, those which were not diagnosed with osteoporosis were selected as controls (n = 8). Taking into account the fact that bed rest may alter the expression of miRs, all the women’s enrolled for the study were questioned about their history of bed rest and it was confirmed that no subject had a 6 month history of being bed ridden. Blood from included volunteers was collected and total RNA were extracted with the help of an RNA extraction kit and were stored at −80 °C for qRT-PCR studies involving the expression of miRs and TRAF6. Prior to the study the volunteers were informed about the purpose of the study and written consent forms were filled from all the subjects before collecting blood samples. The study received approval from the Ethical Review board of Medical University and followed the Declaration of Helsinki for medical protocol and ethics.
Cell Transfection
For in vitro studies we selected MC3T3-E1 cells which were bought from Sigma Aldrich USA. The cells were stored and maintained in MEM which was added to glutamine (4 mM), fetal bovine serum (10%) and penicillin (1%) at room temperature under humid condition with 5%CO2. The cells were added to ascorbic acid (50 mg/L) and β‐glycerophosphate (10 mM) to induce osteogenic differentiation. To inhibit or promote the expression of miR-125b, the cells received transfection of miR-125b mimic or inhibitor for 48 hours with the help of Lipofectamine transfection reagent (Life Techno). To induce the overexpression of TRAF6, to build the overexpressing pcDNA3.1-TRAF6 vector, the cDNA of TRAF6 was cloned in pcDNA3.1 (Invitrogen, USA). The MC3T3-E1 cells were transfected with siRNA-TRAF6 for knock-down expression levels of TRAF6.
Cell Viability and Toxicity Studies
The viability of cells was done by Cell Counting kit-8 assay (CCK-8). The MC3T3-E1 cells were maintained in 96 well plates with 6×103 cells/well; the wells were added to CCK-8 reagent (10 µL) and incubated for 2 hours. The optical density was recorded with the help of microplate reader selecting a wavelength of 450 nm. The cell toxicity was assessed using the LDH-assay kit (Sigma-Aldrich, USA) following the supplied instructions.
Luciferase Reporter Assay
To confirm the predicted target genes of miR-125b by TargetScan, a luciferase assay was performed. The dual reporter vectors (Promega, USA) were cloned with Mutant (Mut) 3ʹUTR or a wild-type (WT) sequence of TRAF6. After this the MC3T3-E1 cells were co-transfected with WT or Mut TRAF6 3ʹUTR plasmid along with miR-125b mimic or negative control (NC).14 After 48 hours of transfecting the cells, the cells were processed to detect the luciferase activity with the help of dual-luciferase reporter assay (Promega, USA).
Rat Model of Ovariectomized Osteoporosis
The rat model of postmenopausal osteoporosis was created by performing ovariectomies on female rats. All the experiments were approved by the Animal Ethical Committee of the First Affiliated Hospital of Anhui Medical University, China (Approval No.00676B). The experiments were in adherence to the ARRIVE guidelines for experiments on animals. The experimental procedures were in agreement with the ‘Regulations for the Administration of Affairs Concerning Experimental Animals’ China, for welfare of experimental animals issued by the government of the Peoples Republic of China under “Laws of the People’s Republic of China”. Sprague-Dawley rats (female) weighing between 260–280 grams were supplied by the animal house of the First Affiliated Hospital of Anhui Medical University, China. The rats were kept in individual polypropylene cages and were exposed to 12 hour dark and light cycle, the temperature was maintained at 25 ± 1 °C with humidity of 50 ± 10%, with free access to pellet diet and water. After normalization period of 7 days, the female rats were distributed randomly into four groups (n = 10). The groups were as follows, Group 1: Ovariectomized rats, Group 2: Sham operated, Group 3: Ovariectomized + miR-125b inhibitor treated, Group 4: Ovariectomized + siRNA-TRAF6. The rats prior to operation were anesthetized with ketamine and xylazine. 30 days after operating the rats were injected with 100 μL of miR-125b inhibitor or siRNA-TRAF6 via subcutaneous route. After 28 days of dosing the rats were submitted to ketamine and xylazine anesthesia and sacrificed. The rats were operated on and left femur bones were removed and analyzed for bone mineral density and other biomechanic parameters. The right femur bones were frozen at −80 °C to estimate the protein and RNA content.
Bone Mineral Density and Biomechanics Analysis
The harvested femurs were submitted to dual-energy X-ray absorptiometry (DEXA) using a PIXImus II densitometer (GE medical systems, USA). The bones were also analyzed for biomechanical parameters which included maximum load bearing capacity, bending stress and elasticity using the mechanical testing machine.
Quantitative-Reverse Transcription-Quantitative Polymerase Chain Reaction (qRT‐PCR)
The total RNAs were separated using TRIzol reagent from the cells and bone tissues. The cDNA was synthesized using the cDNA synthesis kit (ThermoFisher, USA) following the supplied instructions. The RT-qPCR was performed using the ABI-7500 detection system (Applied biosystems, USA). To estimate the relative expression GAPDH and U6 were selected as controls. The primers used for the experiment were obtained from Sangon Biotech (China).
Western Blot Analysis
Total proteins were isolated using RIPA buffer from both cells and bone tissues then analyzed with a protein estimation kit (Sigma Aldrich, USA). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels were used to separate 10 µg of proteins and transferred to PVDF membranes. Prior to incubating the membranes with antibodies they were blocked with non-fat milk (5%). The antibodies used were anti-Runx2 (1:500 Abcam), anti-RANKL (1:500 Abcam), anti-BMP2 (1:500 Abcam), anti-OPG (1:500, Abcam), anti-p-STAT3 (1:500 Abcam) and anti-p-JAK2 (1:500 Abcam). The antibodies were incubated with membranes for 12 hours at 4 °C. The blots were then incubated with secondary antibodies for 1 hour at room temperature and visualized with the aid of chemiluminescent reagent and densitometric analysis was done by image reading software (Bio-Rad, USA). The relative expression of bands was done using actin as a loading control.
Statistics
All the statistics were done using GraphPad Prism software. All the results were depicted as mean ± standard deviation (SD). The variation between the groups was done by one way analysis of variance (ANOVA). The correlation between the groups was done by Pearson analysis. The values of P<0.05 were considered significant.
Results
miR‐125b is Over-Expressed in Postmenopausal Osteoporosis Patients
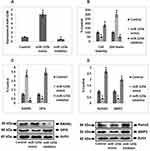
To outline the expression of miR-125b in osteoporosis we screened three miRNA expression datasets from the Omnibus datasets. Three databases were GDS928, GDC1287 and GDS3353 which involved GSE1367, GSE6711 and GSE7429 series, respectively, in them. The downloaded datasets showed that miR-125b is one of the significantly over-expressed miRs in the osteoporosis group compared to the control group (Figure 1A–C). The study of miRNA profiles in postmenopausal subjects with osteoporosis showed similar results (Figure 1D). The findings, hence, suggested that miR-125b is over-expressed in osteoporosis.
miR‐125b Inhibits Cell Viability and Promoted Release of Lactate Dehydrogenase
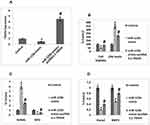
Expression of miR-125b was significantly over-expressed in clinical samples of postmenopausal osteoporosis subjects and also in the Omnibus datasets. This finding prompted us to evaluate the possible biological functioning of miR-125b. The MC3T3-E1 cells were transfected with miR-125b mimic, or inhibitor, separately. RT-qPCR was done to assess the efficiency of transfection (Figure 2A). Cell counting kit-8 (CCK-8) was used, the outcomes indicated that transfection of the miR-125b mimic caused a significant inhibition of cell viability whereas miR-125b inhibition caused increased cell viability. In addition to this, miR-125b mimic caused a significant increase in levels of lactate dehydrogenase compared to the control group (Figure 2B).
miR‐125b Enhanced the Ratio of RANKL/OPG and Suppressed the Expression of Runx2 and BMP2
Literature has confirmed that RANKL/OPG is a crucial marker indicating balance between the process of bone formation and the resorption process. Expression of RANKL and OPG was done by Western blot analysis, the results suggested that expression levels of RANKL were significantly increased in cells transfected with miR-125b mimic, whereas the expression of OPG was suppressed under similar transfection conditions. An inverse outcome for expression of RANKL/OPG ratio was observed in cells transfected with miR-125b inhibitor (Figure 2C). BMP2 and Runx2 are important markers of osteogenic differentiation, the outcomes indicated that their expressions were suppressed in cells treated with miR-125b mimic whereas the transfection of miR-125b inhibitor promoted the expressions of Runx2 and BMP2 significantly in the cells (Figure 2D).
TRAF6 is a Favorable Target Gene of miR‐125b
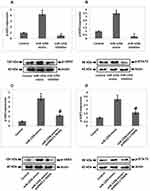
To study the involved mechanism behind miR-125b in osteoporosis, we performed a prediction study for the favorable target genes of miR-125b by TragetScan. We found that from the predicted genes the 3ʹUTR region of TRAF6 had a potential site which was complimentary to the seed region of miR-125b (Figure 3A). Further, luciferase reporter assay suggested that miR-125b mimic caused a significant decrease in the luciferase activity of TRAF6. However, we found no significant changes for the mutant reporter (Figure 3B). Altogether, the outcomes clearly suggested that the TRAF6 transcript was the potential target of miR-125b in MC3T3-E1 cells.
Expression of miR‐125b Negatively Associated with TRAF6
We plotted the expression levels of miR-125b and TRAF6 in osteoporosis samples followed by Pearson correlation analysis and the calculation of a regression coefficient, it was found that expression of miR-125b was negatively linked with TRAF6 with R2 = 0.267, P = 0.0085 (Figure 3C). To further confirm these statistics, i.e. the negative correlation of miR-125b on TRAF6, we performed RT-qPCR on miR-125b mimic or inhibitor transfected MC3T3-e1 cells. The findings (Figure 3D) suggested that miR-125b mimic caused a significant inhibition of TRAF6 levels while miR-125b inhibitor showed the opposing role on the mRNA levels of TRAF6.
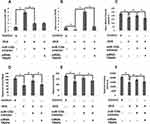
Up-Regulation of TRAF6 Reversed the miR-125b Mediated Cell
To conclude if miR-125b could promote osteoporosis via targeting TRAF6, we transfected the MC3T3-E1 cells with the over-expressing vector of pcDNA3.1-TRAF6 (Figure 4A). We found that when compared with the miR-125b mimic transfected group the over-expression of TRAF6 caused a significant elevation in cell viability and also inhibited the release of lactate dehydrogenase (Figure 4B). We also found that, elevated RANKL/OPG ratio mediated by miR-125b was inverted after transfection of pcDNA3.1-TRAF6 (Figure 4C). Similar outcomes were found in relevant expressions of Runx2 and BMP2 (Figure 4D).
JAK2/STAT3 Axis is Involved in miR-125b-TRAF6 Mediated Osteoporosis
In order to find the involved molecular mechanism for miR-125b and TRAF6 on osteoporosis we evaluated the JAK2/STAT3 signaling pathway, which was the downstream pathway of miR-125b. As evidenced (Figure 5A and B), the cells transfected with miR-125b mimic caused a significant over-expression of pJAK2 and STAT3 compared to the control group cells, whereas transfection of miR-125b inhibitor decreased the levels of pJAK2 and pSTAT3 induced by miR-125b mimic (Figure 5C and D). The outcomes of the experiment suggested that the JAK2/STAT3 cascade is involved in the development of osteoporosis.
The Progression of Osteoporosis in OVX Rats Was Regulated by the miR-125b-TRAF6 Axis
For in vivo studies we successfully developed the OVX model in rats which were used to develop osteoporosis in vivo followed by mRNA profiling to express miR-125b and TRAF6 by RT-qPCR (Figure 6A and B). In the OVX rats we found that the bone mineral density, maximum bending stress, elasticity and maximum load of the femur bones were found to be suppressed significantly compared to sham operated rats. However, these decreased values were reversed in rat’s transfected with miR-125b inhibitor. Further we performed rescue experiments to confirm the involvement of TRAF6 in vivo. The outcomes suggested that bone mineral density and the biomechanical parameters, such as maximum bending stress, elasticity and maximum load, were significantly improved in rats receiving siRNA-TRAF6 (Figure 6C–F). A similar pattern was seen in the evaluation of Runx2 and BMP2 (Figure 7A and B). These outcomes suggested that miR-125b-TRAF6 pathway is responsible for regulating OVX mediated osteoporosis in rats.
For the miR-125b-TRAF6 Axis JAK2/STAT3 Pathway is the Downstream Target
For validating the mechanism of osteoporosis in vivo we studied the JAK2/STAT3 cascade in rats. The outcomes showed that, as compared to sham operated rats, the OVX rats showed increased levels of pJAK2 and pSTAT3, whereas the miR-125b inhibitor decreased the levels of pJAK2 and pSTAT3, suggesting that miR-125 could regulate the JAK2/STAT3 cascade in OVX rats. It was also demonstrated that suppression of TRAF6 restored the levels of pJAK2 and pSTAT3 in OVX rats, indicating that TRAF6 is one of the major links between miR-125b and the JAK2/STAT3 pathway (Figure 7C and D).
Discussion
The current study evidenced that miR-125b was overexpressed in clinical osteoporosis biological samples. It was observed that up-regulated miR-125b significantly promoted the progression of osteoporosis. In the study we also identified TRAF6 as the gene which was found to be the potential target of miR-125b. We encountered a negative regulatory effect between miR-125b and TRAF6 mRNA in subjects who were identified for postmenopausal osteoporosis. Consistent with this, the over-expression of TRAF6 halted the progression of osteoporosis. It was also found that the JAK/STAT3 pathway was activated by miR-125b and TRAF6. All these outcomes were confirmed using the ovariectomized rats.
Prior to this, studies have suggested aberrant expressions of miRs as an inevitable feature of various disorders which include osteoporosis, also miRs are important diagnostic biomarkers in osteoporosis.15 Looking into the earlier studies involved the profiling of miRNAs in osteoporosis, we evidenced miR-125b as a dysregulated miRNA in the 3 databases which we screened and the expression levels of miR-125b in postmenopausal subjects confirmed these results.We first assessed the function of miR-125b in MC3T3-E1 cells, the outcomes indicated that miR-125b inhibited the cell viability and promoted the release of lactate dehydrogenase. The in vivo experiments in rats suggested that miR-125b decreased the bone mineral density and other biomechanical features of femur bones in rats. However, there are multiple factors linked to osteoporosis which include the deficiency of Vitamin D, which features in metabolic disorders related to calcium and phosphate.16 Interestingly, miR-125b has been reported to regulate vitamin D receptors.17 Hence, a possible link between Vitamin D and miR-125b in relevance to osteoporosis should be taken into account in further studies. OPG and RANKL are found to be the important factors which are involved in monitoring the balance between the process of bone formation and resorption.18 RANKL is responsible for initiating the process of bone resorption by enhancing the osteoclastic processes,19 hence an increased ratio of RANKL/OPG could lead to altered bone homeostasis and lead to osteoporosis. Our findings suggested that in MC3T3-E1 cells miR-125b resulted in an increased RANKL/OPG ratio. BMP2 is regarded as important in promoting osteogenic differentiation by triggering Runx2 and, in response, regulating the bone related markers.20 In the present work, miR-125b caused a significant decrease in the expression of BMP2 and Runx2 in rats and in MC3T3-E1 cells, suggesting that miR-125b could inhibit the osteogenic differentiation. We also screened the potential target of miR-125b with the bioinformatics analysis and evidenced that TRAF6 is the downstream target gene for miR-125b. TRAF6 was found to play an important factor, responsible for transcriptional responses in epithelial cells responsible for propagating the type 17 immune responses in inflammatory disorders.21 TRAF6 is found to negatively regulate the JAK/STAT pathway via binding itself to STAT3.22 TRAF6 is also a negative regulator of the cytokine pathway involved in the progression of many inflammatory disorders.23 Many studies have emerged in which TRAF6 have emerged as a potential target of miRNAs such as miR-146a which modulates the immune function by targeting TRAF6,24 miR-124 regulates the expression of TRAF6 in colorectal cancer,25 very importantly a recent report concluded that has-miR-125b-5p acts as a negative regulator MMP-13 via the TRAF6 signaling involving the MAPK/NF-KB cascade in human chondrocytes.26 In light of these reports, we validated that the 3ʹUTR of TRAF6 has a potential binding site for miR-125b by performing a luciferase assay. We also evidenced that the expression of TRAF6 was negatively regulated by miR-125b in the osteoporosis tissues. In addition to this, up-regulation of TRAF6 reversed the effect of miR-125b on the progression of osteoporosis both in MC3T3-E1 cells and ovariectomized rats.
To confirm the mechanism involved by which the miR-125b-TRAF6 axis regulated the progression of osteoporosis, we directed our study towards the JAK/STAT3 pathway, which is reported to be the downstream pathway of TRAF6 in various disorders. It has been reported that the expression of JAK/STAT is important in the progression of various disorders. The JAK2/STAT3 pathway has been evidenced to play an important role in the development and progression of osteoporosis, a study suggested that activation of this pathway encouraged the expression of RANKL and increased the differentiation of osteoclast.27 In a similar way, RANKL has been reported to mediate the osteoclastogenesis via the JAK2/STAT3 pathway.28 In addition to this, the JAK2/STAT3 cascade has been identified to be involved in the regulation of multiple miRs, like miR-30a and miR-630.29,30 In the present study, the results showed that miR-125b mimic enhanced the expression of pJAK2 and p-STAT3 whereas the inhibitor suppressed their levels, indicating the regulatory function of miR-125b on the JAK2/STAT3 pathway. Also the functioning of TRAF6 was confirmed by pcDNA3.1-TRAF6 and siRNATRAF6 in the MC3T3-E1 cells and ovariectomized rats. In line with earlier studies,22,31 we confirmed that TRAF6 is the center of signal transduction between miR-125b and the JAK2/STAT3 cascade in osteoporosis.
In conclusion, miR-125b was evidenced for the first time to be over-expressed in postmenopausal osteoporosis. The work also confirmed that miR-125b encouraged osteoporosis via targeting the TRAF6 gene and involving the JAK2/STAT3 pathway. These findings concluded a novel mechanism underlying the progression of osteoporosis and showed that miR-125b can be a novel therapeutic target for subjects reporting for postmenopausal osteoporosis.
Data Sharing Statement
The data will be available after request to the university.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81702134).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Compston JE, Mcclung MR, Leslie WD. Osteoporosis. Lancet. 2019;393:364–376. doi:10.1016/S0140-6736(18)32112-3
2. Xu X, Jia X, Mo L, et al. Intestinal microbiota: a potential target for the treatment of postmenopausal osteoporosis. Bone Res. 2017;5:17046.
3. Chen X, Zhang S, Chen X, et al. Emodin promotes the osteogenesis of MC3T3‐E1 cells via BMP‐9/Smad pathway and exerts a preventive effect in ovariectomized rats. Acta Biochimica et Biophysica Sinica. 2017;49:867–878.
4. Eastell R, O’Neill T, Hofbauer L, et al. Postmenopausal osteoporosis. Nat Rev Dis Primers. 2016;2:1–16.
5. Black DM, Rosen CJ. Postmenopausal osteoporosis. N Engl J Med. 2016;374:
6. Liu M, Liu L, Bai M, et al. Hypoxia-induced activation of Twist/miR‐214/E‐cadherin axis promotes renal tubular epithelial cell mesenchymal transition and renal fibrosis. Biochem Biophys Res Commun. 2018;495:2324–2330. doi:10.1016/j.bbrc.2017.12.130
7. Wang X, Li G. MicroRNA‐16 functions as a tumor‐suppressor gene in oral squamous cell carcinoma by targeting AKT3 and BCL2L2. J Cell Physiol. 2018;233:9447–9457. doi:10.1002/jcp.26833
8. Zhang X, Zhu Y, Zhang C, et al. miR‐542‐3p prevents ovariectomy‐induced osteoporosis in rats via targeting SFRP1. J Cell Physiol. 2018;233:6798–6806. doi:10.1002/jcp.26430
9. Li X, Ning L, Zhao X, Wan S. MicroRNA‐543 promotes ovariectomy‐induced osteoporosis through inhibition of AKT/p38 MAPK signaling pathway by targeting YAF2. J Cell Biochem. 2019;120:8561–8569.
10. Bellavia D, Salamanna F, Raimondi L, et al. Deregulated miRNAs in osteoporosis: effects in bone metastasis. Cell Mol Life Sci. 2019;76(19):3723–3744. doi:10.1007/s00018-019-03162-w
11. Kozlova A, Pachera E, Maurer B, et al. FRI0402 Microrna-125b regulates apoptosis in systemic sclerosis. Ann Rheum Dis. 2018;77:733–734.
12. Wang X, Ha T, Zou J, et al. MicroRNA-125b protects against myocardial ischaemia/reperfusion injury via targeting p53-mediated apoptotic signalling and TRAF6. Cardiovasc Res. 2014;102(3):385–395. doi:10.1093/cvr/cvu044
13. Jalukar SV, Hostager BS, Bishop GA. Characterization of the roles of TNF receptor-associated factor 6 in CD40-mediated B lymphocyte effector functions. J Immunol. 2000;164:623–630. doi:10.4049/jimmunol.164.2.623
14. Liu X, Xu X. MicroRNA-137 dysregulation predisposes to osteoporotic fracture by impeding ALP activity and expression via suppression of leucine-rich repeat-containing G-protein-coupled receptor 4 expression. Int J Mol Med. 2018;42(2):1026–1033. doi:10.3892/ijmm.2018.3690
15. Pala E, Denkçeken T. Differentially expressed circulating miRNAs in postmenopausal osteoporosis: a meta‐analysis. Biosci Rep. 2019;39:R20190667. doi:10.1042/BSR20190667
16. Bellavia D, Costa V, De Luca A, et al. Vitamin D level between calcium‐phosphorus homeostasis and immune system: new perspective in osteoporosis. Curr Osteoporos Rep. 2016;10:1–12.
17. Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer. 2009;125(6):1328–1333. doi:10.1002/ijc.24459
18. Kondegowda N, Fenutria R, Pollack I, et al. Osteoprotegerin and denosumab stimulate human beta cell proliferation through inhibition of the receptor activator of NF‐κB ligand pathway. Cell Metab. 2015;22:
19. Harper E, Forde H, Davenport C, Rochfort K, Smith D, Cummins P. Vascular calcification in type‐2 diabetes and cardiovascular disease: integrative roles for OPG, RANKL and TRAIL. Vascul Pharmacol. 2016;82:
20. Li J, Hao L, Wu J, Zhang J, Su J. Linarin promotes osteogenic differentiation by activating the BMP‐2/RUNX2 pathway via protein kinase A signaling. Int J Mol Med. 2016;37:
21. Dainichi T, Matsumoto R, Mostafa A, Kabashima K. Immune control by TRAF6-mediated pathways of epithelial cells in the EIME (epithelial immune microenvironment). Front Immunol. 2019;10:1107. doi:10.3389/fimmu.2019.01107
22. Wei J, Yuan Y, Jin C, et al. The ubiquitin ligase TRAF6 negatively regulates the JAK-STAT signaling pathway by binding to STAT3 and mediating its ubiquitination. PLoS One. 2012;7(11):e49567. doi:10.1371/journal.pone.0049567
23. Funakoshi-Tago M, Kamada N, Shimizu T, et al. TRAF6 negatively regulates TNFalpha-induced NF-kappaB activation. Cytokine. 2009;45(2):72–79. doi:10.1016/j.cyto.2008.10.010
24. Walsh MC, Lee J, Choi Y. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol Rev. 2015;266(1):72–92. doi:10.1111/imr.12302
25. Wei C, Lei L, Hui H, Tao Z. MicroRNA-124 regulates TRAF6 expression and functions as an independent prognostic factor in colorectal cancer. Oncol Lett. 2019;18(1):856–863. doi:10.3892/ol.2019.10358
26. Rasheed Z, Rasheed N, Abdulmonem WA. MicroRNA-125b-5p regulates IL-1β induced inflammatory genes via targeting TRAF6-mediated MAPKs and NF-κB signaling in human osteoarthritic chondrocytes. Sci Rep. 2019;9:6882. doi:10.1038/s41598-019-42601-3
27. Zhu J, Tang Y, Wu Q, Ji Y, Feng Z, Kang F. HIF‐1α facilitates osteocyte‐mediated osteoclastogenesis by activating JAK2/STAT3 pathway in vitro. J Cell Physiol. 2019;234(11):21182–21192. doi:10.1002/jcp.28721
28. Li C, Zhao J, Sun L, et al. RANKL downregulates cell surface CXCR6 expression through JAK2/STAT3 signaling pathway during osteoclastogenesis. Biochem Biophys Res Commun. 2012;429:156–162. doi:10.1016/j.bbrc.2012.10.122
29. Yuan F, Chen Y, Zhao Y, et al. microRNA‐30a inhibits the liver cell proliferation and promotes cell apoptosis through the JAK/STAT signaling pathway by targeting SOCS‐1 in rats with sepsis. J Cell Physiol. 2019;234(10):17839–17853. doi:10.1002/jcp.28410
30. Pan X, He X, Yang Y, et al. MiR‐630 inhibits papillary thyroid carcinoma cell growth, metastasis, and epithelial‐mesenchymal transition by suppressing JAK2/STAT3 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:2453.
31. Ma H, Wang X, Ha T, et al. MicroRNA-125b prevents cardiac dysfunction in polymicrobial sepsis by targeting TRAF6-mediated nuclear factor κB activation and p53-mediated apoptotic signaling. J Infect Dis. 2016;214(11):1773–1783. doi:10.1093/infdis/jiw449
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.