Back to Journals » OncoTargets and Therapy » Volume 12
Overexpression of miR-664 is associated with poor overall survival and accelerates cell proliferation, migration and invasion in hepatocellular carcinoma
Authors Wang X, Zhou Z, Zhang T, Wang M, Xu R, Qin S, Zhang S
Received 25 September 2018
Accepted for publication 3 January 2019
Published 28 March 2019 Volume 2019:12 Pages 2373—2381
DOI https://doi.org/10.2147/OTT.S188658
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Xianming Wang,* Zhengtong Zhou,* Tao Zhang,* Minghai Wang, Rongwei Xu, Shiyong Qin, Shuguang Zhang
Department of General Surgery, Qianfoshan Hospital Affiliated to Shandong University, Shandong 250014, China
*These authors contributed equally to this work
Introduction: Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death worldwide. This study aimed to investigate the expression patterns of microRNA-664 (miR-664) in HCC tissues and cells, and assess its clinical significance and functional role in HCC.
Patients and methods: One hundred and thirty-four paired HCC and non-cancerous tissues were collected from patients who underwent surgery in Qianfoshan Hospital affiliated to Shandong University (Shandong, China) between 2009 and 2012. Expression of miR-664 was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Prognostic value of miR-664 in HCC was evaluated using Kaplan–Meier survival analysis and Cox regression analysis. Cell proliferation was analyzed using the CCK-8 assay, and cell migration and invasion of HCC cells was evaluated by the Transwell assay.
Results: Expression of miR-664 was significantly upregulated in HCC tissues and cells when compared with the normal controls (all P<0.05). MiR-664 expression was associated with lymph node metastasis, TNM stage and differentiation (all P<0.05) in the HCC patients. High miR-664 expression predicted poor overall survival (log-rank P=0.004) and acted as an independent prognostic factor (HR =1.945, 95% CI=1.078–3.508, P=0.027). According to cell experiments, the upregulation of miR-664 could promote, whereas the downregulation of miR-664 could inhibit proliferation, migration and invasion of HCC cells (all P<0.05). SIVA1 was predicted as a direct target gene of miR-664 in HCC.
Conclusion: All data indicated that overexpression of miR-664 is associated with poor prognosis of HCC patients, and may enhance tumor progression of HCC by targeting SIVA1. MiR-664 may be a candidate therapeutic target for HCC treatment.
Keywords: MiR-664, prognosis, proliferation, migration, invasion, hepatocellular carcinoma, tumor progression
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies with high rates of incidence and mortality.1 It is the second leading cause of cancer mortality, lending to this, HCC is a serious health burden worldwide.2,3 Researchers have identified common risk factors for HCC occurrence along with the increased incidence rate such as: liver cirrhosis, viral infections and metabolic diseases.4–6 Despite advances in therapeutic methods including: surgery, chemotherapy and radiotherapy, the prognosis and outcomes of patients suffering with HCC are still dismal.7 Thus, more effective therapies are urgently needed to meet the clinical requirements of HCC treatment. In recent studies, targeted therapy has attracted attention in the treatment of various human cancers.8 This therapeutic strategy mainly relies on the identification of molecular targets with obvious clinical and functional roles in disease progression.9,10 Therefore, we considered that it is important to discover novel therapeutic target molecules for HCC.
MicroRNAs (miRNAs) have been highlighted in recent research for their critical roles in tumor initiation and development.11,12 They are a group of small RNAs without the capacity of protein-coding, and have important regulatory functions in gene expression at post-transcriptional levels.13 MiRNAs have been reported to be involved in various biological processes, such as cell proliferation, differentiation, invasion, migration, cell cycle and cell apoptosis, in both normal and abnormal cells, especially tumor cells.14,15 Emerging evidence has indicated that miRNAs could modulate tumor progression by regulating oncogenes or tumor suppressors in different types of human cancer.16 Additionally, they serve as oncogenes or tumor suppressors themselves, and thus are used as therapeutic targets in diverse malignancies.17,18 MicroRNA-664 (miR-664) has been reported to be involved in tumor growth, cell differentiation and apoptosis in some cancers.19,20 A previous study found upregulation of miR-664 in HCC samples compared with normal controls.21 However, the clinical significance and functional role of miR-664 in HCC remains elusive, and warrants in-depth studies.
To explore novel therapeutic targets and further understand the role of miR-664 in HCC, we investigated the expression patterns of miR-664 in HCC samples, assessed its prognostic value, as well as its biological function in tumor progression.
Patients and methods
Patient selection and tissue collection
This study was carried out in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Qianfoshan Hospital affiliated to Shandong University (Shandong, China). Written informed consent was obtained from each patient. HCC and non-cancerous tissue were collected from 134 HCC patients who underwent surgery in the Qianfoshan Hospital affiliated to Shandong University (Shandong, China) between 2009 and 2012. All tissue specimens were evaluated and approved by two experienced pathologists and immediately frozen in liquid nitrogen for RNA extraction. The enrolled patients had not received any preoperative therapy. The clinicopathological characteristics of the patients are summarized in Table 1. After the surgery, patients were followed up for 5 years, and their survival information was recorded for subsequent survival analysis.
  | Table 1 Relationship between miR-664 expression and clinicopathological features of HCC patients |
Cell culture and transfection
Three HCC cell lines, Huh7, Hep3B, MHCC97, and one normal human hepatic cell line LO2 were purchased from the American Type Culture Collection (ATCC). A normal human hepatic cell line LO2 was obtained as negative control. All these cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS, Gibco, CA, USA) added at 37°C in a humidified atmosphere with 5% CO2.
To regulate the expression of miR-664 in HCC cells, cell transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific) as per the manufacturer’s instructions. The vectors used in cell transfection were as follows: miR-664 mimic, miR-664 inhibitor and the corresponding negative controls (mimic NC and inhibitor NC). In addition, the cells transfected with only transfection reagent were set as mock group. All the vectors were obtained from GeneCopoeia (Guangzhou, China).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was obtained from the HCC tissues and cells using TRIzol reagent (Thermo Fisher Scientific), and evaluated by NanoDrop 2000 (Thermo Fisher Scientific). The pure RNA was reverse transcribed into cDNA by a PrimeScript RT reagent kit (TaKaRa, Shiga, Japan). All these experiments were carried out following the manufacturers’ protocols.
Expression levels of miR-664 were measured using qRT-PCR, which was performed using SYBR green I Master Mix kit (Thermo Fisher Scientific) and 7300 Real-Time PCR System (Thermo Fisher Scientific). The relative expression of miR-664 was calculated by 2−ΔΔCt method and normalized to U6. The sequences of the primers were as follows: miR-664 forward: 3′-ACACTCCAGCTGGGTATTCATTTATCCCCAGCC-5′, reverse: 3′-CTCAACTGGTGTCGTGGA-5′; U6, forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse: 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
CCK-8 assay
Cell proliferation of HCC cells was examined using the CCK-8 assay (Dojindo, Kumamoto, Japan) as per the manufacturer’s protocols. The stably transfected cells were seeded into 24-well plates at a cell density of 5×104/mL, and cultured for 24, 48 and 72 hours at 37°C before addition of CCK-8 (5 mg/mL) to the cultures. The wells were maintained at 37°C for a further 1 hour, then examined using a Thermomax microplate reader at an absorbance of 450 nm. This experiment was repeated three times.
Transwell assay
Cell migration and invasion of HCC cells were investigated using the Transwell assay with 8 μm pore size Transwell chambers (Corning Incorporated, Corning, NY, USA). The upper chambers included serum-free DMEM medium and were coated with Matrigel (Corning Incorporated) for invasion analysis. The lower chambers were filled with DMEM supplemented with 10% FBS. After successful transfection, cells were seeded into upper chambers at a cell density of 5×104/mL and cultured at 37°C for 24 hours. The migrated and invaded cells were stained and counted under an inverted microscope (Olympus Corporation, Tokyo, Japan).
Luciferase reporter assay
In order to investigate the molecular mechanisms underlying the role of miR-664 in HCC, the potential targets of miR-664 were predicted using TargetScan. By the prediction, the complementary sequence was found in the 3′-UTR of SIVA1, which represents an apoptosis-inducing factor. In this study, we used luciferase reporter assay to confirm this potential target gene in MHCC97 cells. The firefly luciferase reporter vectors with Renilla luciferase (Promega, Fitchburg, WI, USA) were constructed with the wild-type 3′-UTR, mutant-type 3′-UTR or control vectors. The cells were seeded into 24-well plates and the constructed vectors were co-transfected with the miR-664 mimic or miR-664 inhibitor. After 48 hours incubation, the luciferase activity was measured using a SecrePair Dual-Luciferase Reporter System (Promega).
Statistical analysis
All data were expressed as a mean ± SD and assessed using SPSS 18.0 software (SPSS Inc., Chicago, IL) and GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Differences between two groups were analyzed using Student’s t-test, and comparison between multiple groups was assessed by one-way ANOVA followed by Tukey’s multiple comparison test. Kaplan–Meier survival analysis was performed to plot survival curves based on miR-664 expression in patients with HCC. The prognostic value of miR-664 was confirmed using Cox regression analysis. A P-value <0.05 was considered statistically significant.
Results
Expression of miR-664 in HCC tissues and cells
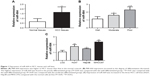
According to qRT-PCR, the expression of miR-664 in HCC tissues was significantly higher than that in the normal controls (P<0.01, Figure 1A). Our research cohort included 48 cases with well differentiated tumors, 44 cases with moderately differentiated tumors and 42 cases with poorly differentiated tumors, and the expression of miR-664 in different differentiation groups was analyzed to preliminarily investigate the role of miR-664 in tumor development. We found that the expression of miR-664 was elevated as the differentiation degree decreases (Figure 1B). The highest expression of miR-664 was detected in the group with poor differentiation (all P<0.05). In addition, the expression of miR-664 in HCC cells was also increased compared to the normal cells (all P<0.05, Figure 1C). And the miR-664 expression was extremely high in Huh7 and MHCC97 (all P<0.01) among the three HCC cell lines.
Relationship between miR-664 expression and clinicopathological features of HCC patients
The research cohort was divided into two subgroups (low/high miR-664 expression group) based on the mean level of miR-664 (1.651). The association of miR-664 expression with clinicopathological characteristics was examined to understand its role in tumor development. The results listed in Table 1 showed that miR-664 expression was associated with lymph node metastasis (P=0.021), TNM stage (P=0.019) and tumor differentiation (P=0.036). The correlation between miR-664 and other clinical factors such as: age, gender, tumor size, cirrhosis and alpha fetoprotein (AFP), was not identified in this analysis (all P>0.05).
Prognostic value of miR-664 in patients with HCC
To evaluate the prognostic value of miR-664 in HCC, the Kaplan–Meier survival curves and Cox analysis were conducted. The survival curves shown in Figure 2 revealed that the patients with high miR-664 levels had a shorter overall survival compared with those with low miR-664 levels (log-rank P=0.004). Furthermore, multivariate Cox analysis indicated that miR-664 (HR=1.945, 95% CI=1.078–3.508, P=0.027) and TNM stage (HR=1.759, 95% CI=1.050–2.948, P=0.032) were two independent prognostic factors (Table 2).
  | Table 2 Multivariate Cox regression analysis for miR-664 in HCC patients |
MiR-664 promotes cell proliferation of HCC cells
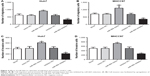
To further understand the functional role of miR-664 in HCC progression, the effects of miR-664 on biological behavior of HCC cells were determined. Cell transfection was performed in Huh7 and MHCC97 cells, which had remarkably high miR-664 expression levels, to modulate the expression of miR-664 in tumor cells. As shown in Figure 3A and B, we found predictably increased miR-664 in the miR-664 mimic group, and markedly decreased miR-664 in the miR-664 inhibitor group compared with the controls (all P<0.01). The tumor cell proliferation measured by CCK-8 assay was promoted by overexpression of miR-664, while was suppressed by knockdown of miR-664 in both Huh7 and MHCC97 cell lines (all P<0.05, Figure 3C and D).
MiR-664 enhances cell migration and invasion of HCC cells
In addition to proliferation, migration and invasion were also assessed under the alteration of miR-664 in HCC cells. We found the cell migration was enhanced in cells with miR-664 overexpression but was inhibited in cells with miR-664 reduction (all P<0.05, Figure 4A and B). Similarly, the upregulation of miR-664 could promote, whereas the downregulation of miR-664 could suppress the capacity of invasion in Huh7 and MHCC97 cells (all P<0.05, Figure 4C and D).
SIVA1 is a target gene of miR-664 in HCC cells
According the target gene prediction, we found that the 3′-UTR of SIVA1 has complementary sequence of miR-664 (Figure 5A). The luciferase reporter analysis was then carried out to confirm this target gene. As shown in Figure 5B, the luciferase activity in the cells with SIVA1 wild-type 3′-UTR (SIVA1-3′-UTR-WT) was remarkably downregulated by the upregulation of miR-664 (P<0.001), while it was upregulated by the knockdown of miR-664 (P<0.001). No effect of miR-664 was observed on the luciferase activity in the cells with SIVA1 mutant-type 3′-UTR (SIVA1-3′-UTR-MT) (all P>0.05).
Discussion
Accumulated evidence has revealed that miRNAs are causally associated with the pathogenesis of various diseases, especially the human cancer.22 They play important roles in tumor initiation and progression in different types of cancers, including HCC. Deng et al reported that miR-29a expression was increased in cholangiocarcinoma tissues and associated with tumor lymph node metastasis, differentiation and clinical stage, indicating the potential oncogenic role of miR-29a in tumor development.23 Another study found that deregulation of miR-584 in thyroid carcinoma could suppress tumor cell migration and invasion by regulation of the oncogene ROCK1.24 In HCC, downregulation of miR-542–3 p has been demonstrated to be involved in the promotion of tumor metastasis and invasion in HCC cells by activating the TGF-β/Smad signaling pathway.25 Another example for miR-342–3 p also indicated the inhibiting effects of miR-342–3 p on cell proliferation and tumor growth in HCC.26 These previous data suggested that it is important to identify functional miRNAs in tumor progression for improvement of cancer treatment.
Dysregulation of miR-664 has been proven to be involved in tumor progression with critical clinical and functional roles. A study by Wu et al27 demonstrated that miR-664 expression was decreased in breast cancer tissues and cells, and inhibited tumor progression in breast cancer cells. Decreased expression of miR-664 has also been reported in cervical cancer samples and was associated with tumor cell growth by regulation of E-Cadherin.28 By contrast, upregulated expression of miR-664 in lung cancer has been reported in a study by Zhu et al,29 which also found that miR-664 overexpression could promote tumor cell proliferation, migration and invasion. Similarly, the expression of miR-664 in osteosarcoma was elevated compared with the normal controls and had facilitated effects on carcinogenesis of this malignancy.30 Given the previous evidence, we considered that the expression of miR-664 may vary with the types of human cancer.
In the present study, we found the expression of miR-664 was significantly upregulated in HCC tissues and cell lines compared to the normal controls. This result was consistent with the previous research by Yang et al,21 which also reported the increased expression of miR-664 in HCC samples. In addition, the expression of miR-664 was found to be increased as the degree of differentiation decreases, indicating that miR-664 may be correlated with tumor development. To further analyze the role of miR-664 in HCC pathogenesis, we assessed the clinical significance and biological function of miR-664 in HCC. According to the chi-squared test, we found that high miR-664 expression was associated with positive lymph node metastasis, advanced TNM stage and poor differentiation of HCC patients, indicating that miR-664 may be involved in aggressive development of HCC. The survival curves plotted using the Kaplan–Meier method revealed that patients with high miR-664 expression had the worst overall survival compared with those with low miR-664 expression. Furthermore, the multivariate Cox analysis indicated that miR-664 was an independent prognostic factor in HCC. Collectively, the upregulated miR-664 expression predicted poor prognosis of HCC.
To understand the role of miR-664 in tumor progression of HCC, the effects of miR-664 expression on proliferation, migration and invasion of HCC cells were consequently investigated. The expression of miR-664 in HCC cells was modulated by cell transfection with the use of miR-664 mimic and miR-664 inhibitor. After the regulation of miR-664, we found that the upregulation of miR-664 enhanced, whereas the downregulation of miR-664 inhibited the capacities of proliferation, migration and invasion of HCC cells. The dysregulation of miR-664 has been previously reported to regulate biological behavior in other tumor cells. In T-cell acute lymphoblastic leukemia cells, miR-664 could promote cell proliferation, migration and invasion by targeting PLP2.31 In osteosarcoma cells, overexpression of miR-664 facilitated cell migration and invasion by downregulation of SOX7.32 Furthermore, we predicted and confirmed that SIVA1 was a direct target of miR-664 in HCC cells. SIVA1 is a pro-apoptotic protein and serves as a critical role in certain apoptosis signaling pathways.33 A previous study by Ma et al34 indicated that SIVA1 could suppress ovarian cancer cell proliferation, migration and invasion by phosphorylating Stathmin. Similarly, Li and his colleagues also found that SIVA1 could inhibit the epithelial-mesenchymal transition in breast cancer cells by regulation of Stathmin.35 Stathmin is also known as oncoprotein 18 and is a microtubule destabilizing protein. It is reported that the phosphorylation and inactivation of Stathmin leads to the suppression of cell proliferation, migration and invasion in various human cancers, including HCC.36,37 Thus, we suspected that the promoting effect on HCC cell proliferation, migration and invasion by miR-664 may be exerted by activating Stathmin via the downregulation of SIVA1. The supposed molecular mechanism is outlined in Figure 6. However, the understanding of the mechanisms underlying the role of miR-664 in HCC remains limited and our hypothesis regarding to the related mechanisms needs to be verified in further studies.
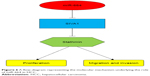  | Figure 6 A flow-diagram representing the molecular mechanism underlying the role of miR-664 in HCC. |
Conclusion
Taken together, all data in our study revealed that overexpression of miR-664 serves as an independent prognostic biomarker in patients with HCC, and promotes proliferation, migration and invasion of HCC cells. These data indicated that miR-664 may be a potential therapeutic target for therapy in HCC.
Acknowledgments
This study was funded by the National Natural Science Foundation (81702752), and Shandong province Natural Science Foundation (ZR2017LH008).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
[No authors listed] Global battle against cancer won’t be won with treatment alone-effective prevention measures urgently needed to prevent cancer crisis. Cent Eur J Public Health. 2014;22(1):28. | ||
An J, Zhang Z, Liu Z, Wang R, Hui D, Jin Y. Overexpression of Cullin7 is associated with hepatocellular carcinoma progression and pathogenesis. BMC Cancer. 2017;17(1):828. | ||
Heidelbaugh JJ, Bruderly M. Cirrhosis and chronic liver failure: part I. Diagnosis and evaluation. Am Fam Physician. 2006;74(5):756–762. | ||
Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13(17):2436–2441. | ||
White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on Systematic review. Clin Gastroenterol Hepatol. 2012;10(12):1342–1359. | ||
Graf D, Vallböhmer D, Knoefel WT, et al. Multimodal treatment of hepatocellular carcinoma. Eur J Intern Med. 2014;25(5):430–437. | ||
Deleon TT, Ahn DH, Bogenberger JM, et al. Novel targeted therapy strategies for biliary tract cancers and hepatocellular carcinoma. Future Oncol. 2018;14(6):553–566. | ||
Ju L, Zhou C. Integrin beta 1 enhances the epithelial-mesenchymal transition in association with gefitinib resistance of non-small cell lung cancer. CBM. 2013;13(5):329–336. | ||
Shaikh MV, Kala M, Nivsarkar M. CD90 a potential cancer stem cell marker and a therapeutic target. CBM. 2016;16(3):301–307. | ||
Chen S, Dai Y, Zhang X, Jin D, Li X, Zhang Y. Increased miR-449a expression in colorectal carcinoma tissues is inversely correlated with serum carcinoembryonic antigen. Oncol Lett. 2014;7(2):568–572. | ||
Zhao X, Zhou Y, Chen YU, Yu F. miR-494 inhibits ovarian cancer cell proliferation and promotes apoptosis by targeting FGFR2. Oncol Lett. 2016;11(6):4245–4251. | ||
Higashi T, Hayashi H, Ishimoto T, et al. miR-9-3p plays a tumour-suppressor role by targeting TAZ (Wwtr1) in hepatocellular carcinoma cells. Br J Cancer. 2015;113(2):252–258. | ||
Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22(1):12. | ||
Sanches JGP, Xu Y, Yabasin IB, et al. miR-501 is upregulated in cervical cancer and promotes cell proliferation, migration and invasion by targeting CYLD. Chem Biol Interact. 2018;285:85–95. | ||
Zhu L, Chen Y, Nie K, Xiao Y, Yu H. MiR-101 inhibits cell proliferation and invasion of pancreatic cancer through targeting STMN1. Cancer Biomark. 2018;23(2):301–309. | ||
Wang C, Li S, Xu J, Niu W, Li S. microRNA-935 is reduced in non-small-cell lung cancer tissue, is linked to poor outcome, and acts on signal transduction mediator E2F7 and the Akt pathway. Br J Biomed Sci. 2019;76(1):17–23. | ||
Gao J, Sr Y, Yuan Y. MicroRNA-590-5p functions as a tumor suppressor in breast cancer conferring inhibitory effects on cell migration, invasion, and epithelial-mesenchymal transition by downregulating the Wnt-beta-catenin signaling pathway. J Cell Physiol. 2019;234(2):1827–1841. | ||
Fiala O, Pitule P, Hosek P, et al. The association of miR-126-3p, miR-126-5p and miR-664-3p expression profiles with outcomes of patients with metastatic colorectal cancer treated with bevacizumab. Tumour Biol. 2017;39(7):1–9. | ||
He JH, Han ZP, Liu JM, et al. Overexpression of long non-coding RNA MEG3 inhibits proliferation of hepatocellular carcinoma Huh7 cells via negative modulation of miRNA-664. J Cell Biochem. 2017;118(11):3713–3721. | ||
Yang H, Cho ME, Li TW, et al. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123(1):285–298. | ||
Chen W, Chu S, Li H, Qiu Y. MicroRNA-146a-5p enhances ginsenoside Rh2-induced anti-proliferation and the apoptosis of the human liver cancer cell line HepG2. Oncol Lett. 2018;16(4):5367–5374. | ||
Deng Y, Chen Y. Increased expression of miR-29a and its prognostic significance in patients with cholangiocarcinoma. Oncol Res Treat. 2017;40(3):128–132. | ||
Xiang J, Wu Y, Li DS, et al. miR-584 suppresses invasion and cell migration of thyroid carcinoma by regulating the target oncogene ROCK1. Oncol Res Treat. 2015;38(9):436–440. | ||
Zhang T, Liu W, Meng W, et al. Downregulation of miR-542-3p promotes cancer metastasis through activating TGF-β/Smad signaling in hepatocellular carcinoma. Onco Targets Ther. 2018;11:1929–1939. | ||
Liu W, Kang L, Han J, et al. miR-342-3p suppresses hepatocellular carcinoma proliferation through inhibition of IGF-1R-mediated Warburg effect. Onco Targets Ther. 2018;11:1643–1653. | ||
Wu L, Li Y, Li J, Ma D. MicroRNA-664 targets insulin receptor substrate 1 to suppress cell proliferation and invasion in breast cancer. Oncol Res. Epub 31 March, 2018. | ||
Yang Y, Liu H, Wang X, Chen L. Up-regulation of microRNA-664 inhibits cell growth and increases cisplatin sensitivity in cervical cancer. Int J Clin Exp Med. 2015;8(10):18123–18129. | ||
Zhu X, Ju S, Yuan F, et al. microRNA-664 enhances proliferation, migration and invasion of lung cancer cells. Exp Ther Med. 2017;13(6):3555–3562. | ||
Chen B, Bao Y, Chen X, et al. Mir-664 promotes osteosarcoma cells proliferation via downregulating of FOXO4. Biomed Pharmacother. 2015;75:1–7. | ||
Zhu H, Miao MH, Ji XQ, Xue J, Shao XJ. miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia. Biochem Biophys Res Commun. 2015;459(2):340–345. | ||
Bao Y, Chen B, Wu Q, et al. Overexpression of miR-664 is associated with enhanced osteosarcoma cell migration and invasion ability via targeting SOX7. Clin Exp Med. 2017;17(1):51–58. | ||
Iorio-Morin C, Germain P, Roy S, Génier S, Labrecque P, Parent JL. Thromboxane A2 modulates cisplatin-induced apoptosis through a Siva1-dependent mechanism. Cell Death Differ. 2012;19(8):1347–1357. | ||
Ma Y, Liu T, Song X, et al. Siva 1 inhibits proliferation, migration and invasion by phosphorylating stathmin in ovarian cancer cells. Oncol Lett. 2017;14(2):1512–1518. | ||
Li N, Jiang P, Du W, et al. Siva1 suppresses epithelial-mesenchymal transition and metastasis of tumor cells by inhibiting stathmin and stabilizing microtubules. Proc Natl Acad Sci Unit States Am. 2011;108(31):12851–12856. | ||
Yuan RH, Jeng YM, Chen HL, et al. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol. 2006;209(4):549–558. | ||
Yurong L, Biaoxue R, Wei L, et al. Stathmin overexpression is associated with growth, invasion and metastasis of lung adenocarcinoma. Oncotarget. 2017;8(16):26000–26012. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.