Back to Journals » International Journal of Women's Health » Volume 14
Overexpression of miR-3653 is Associated with HPV Infection and Serves as a Biomarker in Patients with Cervical Cancer
Authors Cui H, Zhang B, Ruan M, Fang C, Li N, Sun X, Qi J, Zuo R, Zhang S, Rong J
Received 5 January 2022
Accepted for publication 24 May 2022
Published 8 August 2022 Volume 2022:14 Pages 1037—1045
DOI https://doi.org/10.2147/IJWH.S357140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Elie Al-Chaer
Haiyan Cui,1 Baohua Zhang,2 Mei Ruan,3 Chunmei Fang,4 Ning Li,5 Xiaoqin Sun,6 Junmei Qi,1 Rongrong Zuo,1 Shuangshuang Zhang,1 Jiansheng Rong2
1Pathology Department, The Fourth People Hospital of Zibo, Zibo, Shandong, 255067, People’s Republic of China; 2Pathology Department, Zibo Central Hospital, Zibo, Shandong, 255036, People’s Republic of China; 3Oncology Department, The Fourth People Hospital of Zibo, Zibo, Shandong, 255067, People’s Republic of China; 4Gynecology Department, Zibo Lianchi Women & Infants Hospital, Zibo, Shandong, 255000, People’s Republic of China; 5Department of Physical Examination, Zichuan Traditional Chinese Medicine Hospital, Zibo, Shandong, 255100, People’s Republic of China; 6Pathology Department, Zibo Maternal and Child Health Hospital, Zibo, Shandong, 255000, People’s Republic of China
Correspondence: Jiansheng Rong, Pathology Department, Zibo Central Hospital, No. 54 Gongqingtuan West Road, Zibo, Shandong, 255067, People’s Republic of China, Tel/Fax +86 533-2360321, Email [email protected]
Background: Human papillomavirus (HPV) is a major cause of cervical cancer (CC) occurrence. This study aimed to explore whether abnormal microRNA (miR)-3653 is associated with HPV infection and to investigate the clinical value of miR-3653 in the diagnosis and prognosis of CC.
Methods: Tumor tissues and adjacent non-cancerous tissues were collected from 136 patients with CC. Cervical tissues from 101 patients with uterine fibroids were collected as controls. The expression of miR-3653 was measured by quantitative real-time PCR. The ability of miR-3653 to discriminate between HPV positive (HPV+) and HPV negative (HPV-) CC patients, and to discriminate patients from controls was assessed by receiver operating characteristic analysis. Kaplan–Meier curves and Log rank tests were used to evaluate the relationship of miR-3653 with survival of CC patient. Whether miR-3653 could function as a prognostic indicator was evaluated by univariate and multivariate Cox analyses.
Results: miR-3653, highly expressed in CC tissues, was associated with HPV infection, tumor diameter, International Federation of Gynecology and Obstetrics (FIGO) stage and lymph node metastasis in CC patients. Additionally, miR-3653 was increased in HPV+ controls, CC patients and CC cells. Moreover, miR-3653 could screen HPV+ controls, screen HPV+ patients and screen CC patients. Furthermore, miR-3653 was associated with the survival of CC patients (log-rank P < 0.001) and could serve as an independent prognostic indicator for CC patients.
Conclusion: miR-3653, increased in CC, is related to HPV infection and may serve as a diagnostic and prognostic biomarker for CC patients.
Keywords: miR-3653, microRNA, HPV, cervical cancer, diagnosis, prognosis
Introduction
Cervical cancer (CC), as a prevalent gynecological malignancy, has the second highest mortality rate among other cancers in women worldwide.1,2 Standard primary therapies for CC include radical hysterectomy with pelvic lymphadenectomy, radiotherapy (RT), or a combination of RT and platinum-based chemotherapy.3 Current medical diagnosis and treatment of CC are constantly evolving, but the prognosis of CC is still not optimistic. Human papillomavirus (HPV) infection is known to be a key cause of CC and is closely related to the occurrence and development of CC.4,5 Therefore, exploring the key molecules associated with HPV infection may be expected to provide new potential biomarkers and therapeutic targets for CC.
microRNAs (miRs), a group of small non coding RNAs, are known to be involved in transcriptional and epigenetic regulation through binding to the 3’-untranslated regions (3’-UTRs) of target genes.6 Currently, some functional miRs have been found to be associated with HPV infection in CC, such as miR-21 and miR-93-5p.7,8 In addition, an increasing number of studies have suggested the role of aberrant miRs as biomarkers in various cancers, including CC.9–12 Notably, an earlier study on miRNA expression profiles in HPV infected CC patients has found that the expression levels of miR-3653 in HPV infected CC tissues were significantly higher than that in normal cervical tissues without HPV infection.13 However, the relationship between aberrant expression of miR-3653 and HPV infection is unclear. The differential expression of miR-3653 has been reported in several malignant tumors, including colon cancer, glioma and hepatocellular carcinoma, and the clinical value and biological function of miR-3653 were investigated in these cancers.14–16 Thus, it is significant to explore the role of miR-3653 in CC.
The purpose of this study is to investigate the expression of miR-3653 in CC patients, to explore the correlation between aberrantly expressed miR-3653 and HPV infection, and further evaluate its clinical significance for screening CC patients, as well as its application value for predicting the clinical prognosis of patients with CC. This study will provide a novel biomarker for the diagnosis and treatment of CC patients.
Materials and Methods
Patients and Tissue Collection
The present study was approved by the Ethics Committee of The Fourth People Hospital of Zibo and each participant has provided informed consent. A total of 136 CC patients treated at The Fourth People Hospital of Zibo between 2014 and 2016 were recruited. CC tissues and adjacent non-cancerous tissues were collected from the CC patients. The inclusion criteria of patients were: 1) not receiving chemotherapy, radiotherapy or other adjuvant therapy before the diagnosis of CC was confirmed in patients; 2) they were diagnosed as CC on the pathological basis; and 3) their information had been obtained. The exclusion criteria were as follows: 1) patients were combined with other malignancies, and 2) patients were pregnant or lactating. In addition, we included cervical tissues from 101 patients who underwent hysterectomy due to uterine fibroids and had no cervical lesions or history of malignancy during the same period as the controls. The obtained tissues were frozen and stored in liquid nitrogen for further use. The status of HPV infection was evaluated using HPV-DNA tests. Total DNA was isolated from the tissue samples, and HPV DNA was amplified by PCR using primers MY09/MY11, GP5+/6+ and SPF1/2 (GenePharma, Shanghai, China). The HPV genotypes were further assessed with HPV-type specific PCR.17 Based on the test results, CC patients were divided into 96 HPV positive (HPV+) (64 cases with HPV16, 56 cases with HPV18, and 31 cases with HPV16 & HPV18) and 40 HPV negative (HPV-) patients, and controls were divided into 59 HPV+ (43 cases with HPV16, 33 cases with HPV18, and 18 cases with HPV16 & HPV18) and 42 HPV- cases. All patients participated in 5-year survival follow-up, with death or loss to follow-up as the endpoint.
Cell Culture
HPV- normal immortalized epithelial cell line HaCaT, HPV- CC cell line C33A, HPV16+ CC cell line SiHa and HPV18+ CC cell line HeLa were purchased from the American Type Culture Collection (ATCC), cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS) and placed in humid environment with 5% CO2 and 37°C.
RNA Extraction
Total RNA was extracted from the tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. Then, RNA purity and concentration were detected by NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols.
Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were reverse transcribed into cDNA using a Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, qPCR was performed to measure the expression of miR-3653 using a SYBR Green PCR Kit (Bio-Rad, Shanghai, China) and an Applied Biosystems 7900 Real-Time PCR system (CA, USA). Following were the reaction details: initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15s, 60°C for 20s, and 72°C for 30s. The primers used for qPCR were all synthesized from GenePharma (Shanghai, China), and were as follows: miR-3653 forward, 5’-GCCGAGCTAAGAAGTTGACT-3’ and reverse, 5’-CTCAACTGGTGTCGTGGA-3’; U6 forward, 5’-CTCGCTTCGGCAGCACA-3’ and reverse, 5’-AACGCTTCACGAATTTGCGT-3’. The expression levels of miR-3653 were quantified using the 2−ΔΔCt method,18 and normalized to U6.
Statistical Analysis
The analysis results of quantitative data were presented as mean ± SD. SPSS 22.0 (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad Software, Inc.) was used for statistical analyses, and all experiments were repeated three times. Differences in quantitative data between two groups were compared by Student’s t-test. One-way ANOVA followed by Tukey’s test was used to compare the differences of quantitative data among multiple groups. Chi-square test was used to compare the differences between categorical variables. Receiver operating characteristic (ROC) analysis was used for the diagnostic evaluation of miR-3653 (HPV+ controls vs HPV- controls, HPV+ patients vs HPV- patients, or controls vs CC patients). The relationship between miR-3653 and survival prognosis of CC patients was examined by Kaplan–Meier survival analysis and Log rank test. Univariate and multivariate Cox regression analyses were used to confirm the prognostic value of miR-3653 in CC patients. Differences between groups were considered statistically significant at P < 0.05.
Results
Baseline Features of the Participants
The baseline features of all participants are presented in Table 1. No significant differences were observed between controls and CC patients in age and HPV infection (all P > 0.05). Besides, the classification of CC patients according to tumor diameter, International Federation of Gynecology and Obstetrics (FIGO) stage, histological type, lymph node metastasis and vascular invasion has also been shown in Table 1.
 |
Table 1 Baseline Features of the Participants |
Expression of miR-3653 in CC Patients and Adjacent Non-Cancerous Tissues
Figure 1 demonstrates that miR-3653 expression was significantly increased in CC patients compared with that in adjacent non-cancerous tissues (P < 0.05).
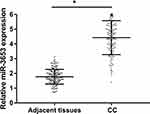 |
Figure 1 Expression of miR-3653 in CC patients and adjacent non-cancerous tissues. *P < 0.05 vs adjacent non-cancerous tissues. Abbreviation: CC, cervical cancer. |
Relationship Between miR-3653 Expression and Clinicopathological Characteristics in CC Patients
The median expression value of miR-3653 (4.44) was used as the cutoff values to classify the patients into low and high miR-3653 expression groups. The results presented in Table 2 reveals that miR-3653 expression was associated with HPV infection (P=0.013), tumor diameter (P=0.036), FIGO stage (P=0.022) and lymph node metastasis (P=0.006) in patients with CC. Besides, there was no association of miR-3653 expression with age, histological type and vascular invasion (all P > 0.05).
 |
Table 2 Relationship Between miR-3653 and Clinicopathological Characteristics of CC Patients |
Expression of miR-3653 in CC Patients and Cell Lines with Different HPV Infection Conditions
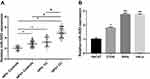
As shown in Figure 2A, after grouping according to HPV infection, miR-3653 expression in HPV+ controls was found to be significantly higher than that in HPV- controls (P < 0.05). In addition, HPV- CC patients had significantly upregulated levels of miR-3653 compared with HPV- control group (P < 0.05). Moreover, we found that HPV+ CC patients had the highest miR-3653 expression, and the difference in miR-3653 expression between HPV+ and HPV- CC patients also reached statistical levels (all P < 0.05). Results of miR-3653 expression levels in cells (Figure 2B) revealed a significant increase of miR-3653 in CC cells compared to that in normal cells; and miR-3653 was higher in both HPV+ CC cells than that in HPV- CC cells (all P < 0.05).
The Ability of Aberrant miR-3653 to Screen HPV+ Controls, HPV+ CC Patients and CC Patients
ROC analysis results demonstrated that miR-3653 was able to differentiate between HPV+ controls and HPV- controls with an area under the curve (AUC) of 0.860 (Figure 3A), and to discriminate between HPV+ and HPV- CC patients with an AUC of 0.902 (Figure 3B). Figure 3C demonstrates that miR-3653 had high diagnostic accuracy in screening CC patients from normal controls. At the cut-off value of 3.445, the AUC, sensitivity and specificity were 0.915, 80.9% and 91.1%, respectively.
Prognostic Value of miR-3653 in Patients with CC
The results of 5-year follow-up data analysis of the present study showed that CC patients with high levels of miR-3653 had a poor prognosis for survival (Figure 4, log-rank P < 0.001). Then, the Cox regression analysis results are presented in Table 3. The results of univariate Cox analysis revealed that FIGO stage, lymph node metastasis, vascular invasion and miR-3653 were significant factors associated with overall survival of CC patients. Then, significant baseline variables from univariate Cox analysis were used for multivariate Cox analysis. We found that FIGO stage [hazard ratio (HR)=1.981, 95% confidence interval (CI)=1.109–3.538, P=0.021] and miR-3653 (HR = 2.447, 95% CI = 1.293–4.629, P=0.006) were independently associated with the overall survival of CC patients.
 |
Table 3 Cox Regression Analysis for Patients with CC |
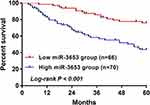 |
Figure 4 High miR-3653 was associated with poor prognosis in CC patients (log-rank P < 0.001). Abbreviation: CC, cervical cancer. |
Discussion
Accumulating evidence has suggested that abnormal expression of miRs is involved in the progression and development of CC. For instance, miR-218, decreased in CC tissues, may play a tumor inhibitory role in CC.1,19 The inhibitory effect of miR-181a on the development of CC has been demonstrated by Luo et al.20 Sun et al have reported that miR-93-5p, which is highly expressed in CC tissues and cells, promotes CC.21 In the present study, we found that miR-3653 was markedly higher in tumor tissues of CC patients than that in adjacent non-cancerous tissues of CC patients. Additionally, the results of association analysis of miR-3653 with clinicopathological characteristics of CC patients indicated that miR-3653 expression was related to HPV infection, tumor diameter, FIGO stage and lymph node metastasis in CC patients. Notably, miR-3653 expression has been reported to be involved in the progression of other types of cancers, such as glioma,15 colon cancer14 and hepatocellular carcinoma.16 Therefore, aberrant miR-3653 expression may be involved in the progression of CC.
High risk (HR)-HPV infection is found to be a main cause of CC. In addition, it is worth noting that some miRs have been shown to be associated with HPV infection. For instance, Liu et al have found the inverse correlation between HPV positivity and miR-218 expression in patients with CC.19 A study by Li et al has reported that miR-93-5p is positively associated with HR-HPV infection.8 miR-3156-3p has been demonstrated to be related to HR-HPV infection.22 In the present study, miR-3653 expression was found to be correlated with HPV infection in CC patients by the Chi-square test. Then, miR-3653 was upregulated in HPV+ controls, CC patients and CC cells compared with that in the corresponding HPV- controls, CC patients and CC cells. In addition, miR-3653 had the ability to discriminate between HPV- and HPV+ controls, and discriminate between HPV- and HPV+CC patients. Therefore, miR-3653 is associated with the HPV infection in CC patients.
It has been known that miRs can function as clinical biomarkers for various cancers. For example, miR-1236-3p expression may serve as a novel biomarker for the diagnosis and prognosis of gastric cancer.23 miR-4730 can serve as a diagnostic and prognostic biomarker for pancreatic cancer.24 Qiu et al demonstrate the role of miR-146a and miR-146b as diagnostic and prognostic biomarkers for papillary thyroid carcinoma.25 More importantly, many miRs have been demonstrated to be used as diagnostic and prognostic biomarkers for CC, such as miR-34a and miR-218,26 miR-314227 and miR-152.28
In the present study, aberrant miR-3653 was suggested to have high diagnostic accuracy in distinguishing CC patients from normal controls. The results of Kaplan–Meier survival analysis demonstrated the significant correlation of miR-3653 with CC patient survival. Besides, Cox analysis results demonstrated that miR-3653 was independently associated with survival prognosis in CC patients. Notably, previous studies have indicated the clinical value of miR-3653 in other cancers. For instance, miR-3653 has been found to function as a prognostic indicator in glioma.15 A study by Lin et al has shown that miR-3653 is an independent marker for predicting the overall survival prognosis in lung adenocarcinoma.29 Low miR‑3653 level is related to poor prognosis in HCC patients.16 Therefore, miR-3653 may be used as a diagnostic and prognostic biomarker for CC patients.
However, this study has some limitations. At first, the sample size was small; thus, a larger sample size is needed in future studies. Second, the mechanism was not investigated. However, miR-3653 has been shown to play an important role in colon cancer via targeting Zeb2,14 and to regulate the progression of hepatocellular carcinoma through repressing integrin‑β1 (ITGB1).16 Therefore, we supposed that miR-3653 may be involved in CC progression by targeting Zeb2 or ITGB1, which needs to be validated in further studies. Third, this study did not include the population with precancerous lesions. The analysis results of this population, especially the HPV-positive group, may provide evidence for miR-3653 to confirm that whether it was associated with tumor progression in HPV-positive women. Therefore, further studies should not only expand the sample size, but also enrich the types of sample source.
In conclusion, this study indicates that increased miR-3653 in CC is closely associated with HPV infection in CC patients. In addition, the differential expression of miR-3653 between CC tumor tissues and normal controls, and its association with overall survival, indicating that miR-3653 may provide a candidate for the development of biomarkers for CC diagnosis and prognosis.
Ethics Statement and Consent to Participate
The present study was approved by the Ethics Committee of The Fourth People Hospital of Zibo and each participant provided informed consent. This study complies with the Declaration of Helsinki.
Disclosure
The authors declare that they have no competing interests.
References
1. Zhu L, Tu H, Liang Y, Tang D. MiR-218 produces anti-tumor effects on cervical cancer cells in vitro. World J Surg Oncol. 2018;16(1):204. doi:10.1186/s12957-018-1506-3
2. Dukic A, Lulic L, Thomas M, et al. HPV oncoproteins and the ubiquitin proteasome system: a signature of malignancy? Pathogens. 2020;9(2):133. doi:10.3390/pathogens9020133
3. Nuchpramool P, Hanprasertpong J. Preoperative neutrophil-lymphocyte ratio and platelet-lymphocyte ratio are not clinically useful in predicting prognosis in early stage cervical cancer. Surg Res Pract. 2018;2018:9162921. doi:10.1155/2018/9162921
4. Zhu L, Chen R, Jiang C, et al. Mechanism underlying long noncoding RNA ILF3AS1mediated inhibition of cervical cancer cell proliferation, invasion and migration, and promotion of apoptosis. Mol Med Rep. 2021;24(2):554. doi:10.3892/mmr.2021.12193
5. Park S, Eom K, Kim J, et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. 2017;17(1):658. doi:10.1186/s12885-017-3642-5
6. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci. 2020;21(5):1723. doi:10.3390/ijms21051723
7. Zamani S, Sohrabi A, Hosseini SM, Rahnamaye-Farzami M, Akbari A. Deregulation of miR-21 and miR-29a in cervical cancer related to HPV Infection. Microrna. 2019;8(2):110–115. doi:10.2174/2211536607666181017124349
8. Li J, Chu ZP, Han H, et al. Suppression of miR-93-5p inhibits high-risk HPV-positive cervical cancer progression via targeting of BTG3. Hum Cell. 2019;32(2):160–171. doi:10.1007/s13577-018-00225-1
9. Sun J, Sun Z, Gareev I, et al. Exosomal miR-2276-5p in plasma is a potential diagnostic and prognostic biomarker in glioma. Front Cell Dev Biol. 2021;9:671202. doi:10.3389/fcell.2021.671202
10. Han L, Sun Y, Lu C, Ma C, Shi J, Sun D. MiR-3614-5p is a potential novel biomarker for colorectal cancer. Front Genet. 2021;12:666833. doi:10.3389/fgene.2021.666833
11. Aftab M, Poojary SS, Seshan V, et al. Urine miRNA signature as a potential non-invasive diagnostic and prognostic biomarker in cervical cancer. Sci Rep. 2021;11(1):10323. doi:10.1038/s41598-021-89388-w
12. Ocadiz-Delgado R, Cruz-Colin JL, Alvarez-Rios E, et al. Expression of miR-34a and miR-15b during the progression of cervical cancer in a murine model expressing the HPV16 E7 oncoprotein. J Physiol Biochem. 2021;77(4):547–555. doi:10.1007/s13105-021-00818-9
13. Gao D, Zhang Y, Zhu M, Liu S, Wang X, Tornesello ML. miRNA expression profiles of HPV-infected patients with cervical cancer in the Uyghur population in China. PLoS One. 2016;11(10):e0164701. doi:10.1371/journal.pone.0164701
14. Zhu W, Luo X, Fu H, Liu L, Sun P, Wang Z. MiR-3653 inhibits the metastasis and epithelial-mesenchymal transition of colon cancer by targeting Zeb2. Pathol Res Pract. 2019;215(10):152577. doi:10.1016/j.prp.2019.152577
15. Chen Y, Li ZH, Liu X, Liu GX, Yang HM, Wu PF. Reduced expression of miR-3653 in glioma and its correlations with clinical progression and patient survival. Eur Rev Med Pharmacol Sci. 2019;23(15):6596–6601. doi:10.26355/eurrev_201908_18547
16. Zhang L, Zhang T, Deng Z, Sun L. MicroRNA3653 inhibits the growth and metastasis of hepatocellular carcinoma by inhibiting ITGB1. Oncol Rep. 2019;41(3):1669–1677. doi:10.3892/or.2019.6971
17. Lee SH, Vigliotti VS, Vigliotti JS, Pappu S. Validation of human papillomavirus genotyping by signature DNA sequence analysis. BMC Clin Pathol. 2009;9:3. doi:10.1186/1472-6890-9-3
18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
19. Liu Z, Mao L, Wang L, Zhang H, Hu X. miR218 functions as a tumor suppressor gene in cervical cancer. Mol Med Rep. 2020;21(1):209–219. doi:10.3892/mmr.2019.10809
20. Luo C, Qiu J. miR-181a inhibits cervical cancer development via downregulating GRP78. Oncol Res. 2017;25(8):1341–1348. doi:10.3727/096504017X14867268787969
21. Sun XY, Han XM, Zhao XL, Cheng XM, Zhang Y. MiR-93-5p promotes cervical cancer progression by targeting THBS2/MMPS signal pathway. Eur Rev Med Pharmacol Sci. 2019;23(12):5113–5121. doi:10.26355/eurrev_201906_18175
22. Xia YF, Pei GH, Wang N, et al. miR-3156-3p is downregulated in HPV-positive cervical cancer and performs as a tumor-suppressive miRNA. Virol J. 2017;14(1):20. doi:10.1186/s12985-017-0695-7
23. An JX, Ma ZS, Ma MH, Shao S, Cao FL, Dai DQ. MiR-1236-3p serves as a new diagnostic and prognostic biomarker for gastric cancer. Cancer Biomark. 2019;25(2):127–132. doi:10.3233/CBM-171026
24. Yang W, Ju HY, Tian XF. Hsa-miR-4730 as a new and potential diagnostic and prognostic indicators for pancreatic cancer. Eur Rev Med Pharmacol Sci. 2020;24(17):8801–8811. doi:10.26355/eurrev_202009_22819
25. Qiu Z, Li H, Wang J, Sun C. miR-146a and miR-146b in the diagnosis and prognosis of papillary thyroid carcinoma. Oncol Rep. 2017;38(5):2735–2740. doi:10.3892/or.2017.5994
26. Wang P, Zhai G, Bai Y. Values of miR-34a and miR-218 expression in the diagnosis of cervical cancer and the prediction of prognosis. Oncol Lett. 2018;15(3):3580–3585. doi:10.3892/ol.2018.7791
27. Luo Q, Wang H, Li J. Serum miR-3142 could be used as a potential biomarker to screen cervical cancer patients from healthy controls. Clin Lab. 2019;65(4). doi:10.7754/Clin.Lab.2018.180925
28. Yang D, Zhang Q. miR-152 may function as an early diagnostic and prognostic biomarker in patients with cervical intraepithelial neoplasia and patients with cervical cancer. Oncol Lett. 2019;17(6):5693–5698. doi:10.3892/ol.2019.10233
29. Lin K, Xu T, He BS, et al. MicroRNA expression profiles predict progression and clinical outcome in lung adenocarcinoma. Onco Targets Ther. 2016;9:5679–5692. doi:10.2147/OTT.S111241
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.