Back to Journals » Cancer Management and Research » Volume 10
Overexpression of lncRNA PANDAR predicts adverse prognosis in acute myeloid leukemia
Authors Yang L, Zhou JD , Zhang TJ, Ma JC, Xiao GF, Chen Q, Deng ZQ , Lin J, Qian J, Yao DM
Received 15 July 2018
Accepted for publication 11 September 2018
Published 29 October 2018 Volume 2018:10 Pages 4999—5007
DOI https://doi.org/10.2147/CMAR.S180150
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lu-Zhe Sun
Lan Yang,1,2,* Jing-Dong Zhou,2,3,* Ting-Juan Zhang,2,3 Ji-Chun Ma,1,2 Gao-Fei Xiao,1,2 Qin Chen,1,2 Zhao-Qun Deng,1,2 Jiang Lin,1,2 Jun Qian,2,3 Dong-Ming Yao1,2
1Laboratory Center, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China; 2The Key Laboratory of Precision Diagnosis and Treatment of Zhenjiang City, Zhenjiang, Jiangsu, People’s Republic of China; 3Department of Hematology, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Background and purpose: Abundant studies have shown that lncRNA PANDAR plays an oncogenic role in human solid tumors. Although abnormal expression of PANDAR has been well investigated in solid tumors, it was rarely studied in hematologic diseases. Hence, the aim of this study was to determine the PANDAR expression level and its clinical significance in patients with acute myeloid leukemia (AML).
Materials and methods: For detecting the expression level of PANDAR in 119 AML patients and 26 controls, real-time quantitative PCR was used in this study. The prognostic values were evaluated by using Kaplan–Meier analysis, Cox regression analyses, and logistic regression analysis.
Results: PANDAR was significantly overexpressed in AML and might be a promising biomarker which could distinguish AML from normal samples (P<0.001). Patients with high expression of PANDAR (PANDARhigh) were older and showed higher bone marrow blasts than patients in PANDARlow group (P=0.029 and 0.032, respectively). Significant differences between these groups were also detected regarding risk group and karyotype finding (P=0.009 and 0.041, respectively). Importantly, PANDARhigh patients presented a significant lower complete remission rate compared to PANDARlow patients (P<0.001). Furthermore, Kaplan–Meier analysis showed that PANDARhigh patients had shorter overall survival compared to PANDARlow patients observing the whole AML cohort, and also in the non-M3 group of patients (P<0.001 and P=0.005, respectively). Multivariate analysis of Cox and logistic regression analysis confirmed that high PANDAR expression was an independent unfavorable risk factor for overall survival and complete remission in both observed patient groups.
Conclusion: These results revealed that PANDAR was overexpressed in AML, and that higher PANDAR expression was associated with poor clinical outcome. Our study therefore suggests that PANDAR expression is a promising biomarker for prognostic prediction for AML.
Keywords: long noncoding RNA, PANDAR expression, acute myeloid leukemia, complete remission, overall survival
Introduction
Acute myeloid leukemia (AML) is a cytogenetically and molecularly heterogeneous disease which is marked by uncontrolled clonal expansion of blast cells.1 Although the new treatment strategies based on molecular biology of AML have been adopted in recent years, the prognosis of the disease remains poor.2–4 It has become apparent that karyotype abnormalities have important value for AML diagnosis classification, prognostic evaluation, and guiding individual treatment.5,6 Cytogenetic aberrations together with several gene mutations including NPM1, CEBPA, TP53, TET2, DNMT3A, and FLT3-ITD have a strong impact on clinical outcome of AML patients.7 In addition to genetic abnormalities, the aberrant expression of some genes, such as overexpression of ERG, BAALC, and EVI1, also has been proven to affect prognosis for AML patients.7 These important findings open up a new field for discovering novel promising biomarkers for AML patients, especially for those who are at risk of poor outcome, so that these patients can be treated with optimized treatment strategies.
Long noncoding RNAs (lncRNAs) are regarded as a kind of noncoding RNA, which are longer than 200 nucleotides. Recently, many studies have reported that lncRNAs play vital roles in gene expression regulation through association with key transcription factors and microRNAs.8,9 lncRNAs could act as an important component in every step of cell biology, which includes the adjustments of transcription initiation and transcription and posttranscriptional level.10 Recently, increasing number of research papers revealed that lncRNAs were relevant to many human diseases, especially to human cancers, and many studies began to explore the molecular mechanisms of lncRNA function in the pathogenesis of these disease or cancers.11 With the deepening of the research, it is becoming increasingly apparent that most of the susceptibility to cancer is not caused by the variation of coding sequences of DNA but by the noncoding regulatory sequences, especially by lncRNAs.10
lncRNA PANDAR, which is located at 6p21.2, plays a vital role in regulation of apoptosis by inhibiting the expression of proapoptotic genes through interaction with the transcription factor NF-YA.12 To date, the abnormal expression of PANDAR has been reported in various solid cancers, such as hepatocellular carcinoma, gastric cancer, and breast cancer.13 However, there are few reports about the expression of PANDAR in blood cancer. Therefore, we focused on exploring the PANDAR expression level and its connection with clinical implication in AML patients.
Materials and methods
Patients and treatment
A total of 119 de novo AML patients and 26 healthy donors were included in the present research, which was approved by the Ethics Committee and Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University. Bone marrow (BM) was collected from all the participants after they signed the informed consents. BM mononuclear cells (BMMNCs) were extracted from BM specimen using Lymphocyte Separation Medium (TBD Sciences, Tianjin, People’s Republic of China). Treatment protocols for AML were described previously.14
Cytogenetics and mutation analysis
By conventional R-banding method, karyotype was analyzed at the time of initial diagnosis. Risk classification based on the karyotype findings has been done as previously reported.15 Mutations in C-KIT, NPM1, DNMT3A, N/K-RAS, and U2AF1 were detected by high-resolution melting analysis,16–20 whereas FLT3-ITD and CEBPA mutations were detected by direct DNA sequencing.21,22
RNA isolation and reverse transcription
Total RNA was extracted by using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The specific procedure of reverse transcription was conducted as previously reported.23
Real-time quantitative PCR
The primers for PANDAR are as follows: forward: 5′-CTCCATCATGCCAA GTTCTGC-3′ and reverse: 5′-GAAGGCAGGCAAGACTCGAA-3′. PANDAR expression was detected by real-time quantitative PCR using AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA). The reaction condition of real-time quantitative PCR was conducted as reported earlier.24,25 Relative PANDAR expression levels were calculated by 2–ΔΔCT method.
Statistical analysis
SPSS software version 20.0 (IBM Corporation, Armonk, NY, USA) was used to carry out the statistical analysis. Meanwhile, receiver operating characteristic (ROC) curve and area under the ROC were applied to assess the value of PANDAR expression. Besides, Pearson’s chi-squared analysis was conducted to detect the difference of categorical variables between PANDARhigh group and PANDARlow group. Through Kaplan–Meier method and Cox regression analysis, the effect of PANDAR expression on prognosis was analyzed. Logistic regression analysis was used to identify the independent risk factors of complete remission (CR). In all tests, P<0.05 was defined as statistically significant.
Results
PANDAR expression in AML
The expression level of PANDAR in controls ranged from 0.000 to 2.926 (median 0.294). PANDAR transcript level in AML patients ranged from 0.005 to 306.109 (median 1.862). Through nonparametric test, PANDAR was found to be significantly upregulated in AML (P<0.001, Figure 1). Besides this, significant upregulation of PANDAR was also found in non-M3-AML and cytogenetically normal AML subgroup of patients (Figure 1).
Distinguishing capacity of PANDAR expression
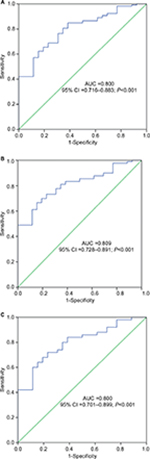
The ROC curve analysis was applied to evaluate whether PANDAR expression could be used as a biomarker for the diagnosis of AML. The results showed that area under the curve value was 0.800 (95% CI: 0.716–0.883), which suggested the PANDAR expression level might be a potential biomarker in discriminating AML from controls (P<0.001, Figure 2A). In addition, when the cutoff value was 0.840, the sensitivity and specificity of diagnosis of AML were 65.5% and 80.8%. For non-M3-AML and CN-AML patients, significant differences also existed (Figure 2B and C, respectively).
The connection between PANDAR expression level and clinical characteristics in AML
By the set cutoff value based on the basis of ROC curve, the whole cohort of AML patients was divided into two groups. Clinical features and laboratory parameters representation between PANDARhigh and PANDARlow groups is separately shown in Table 1. No significant differences were observed in sex, white blood cells (WBCs), hemoglobin, and platelets between two groups (P>0.05). However, patients with PANDAR high expression were older than patients in the PANDAR low-expressed group (P=0.029). Patients in PANDARhigh group showed higher BM blasts than patients in PANDARlow group (P=0.032). Moreover, significant differences between these two groups were also detected regarding risk group and karyotype finding (P=0.009 and 0.041, respectively). Patients in PANDARhigh group had higher frequency of poor karyotypes (15%, 12/78) than patients in PANDARlow group (2%, 1/41). There was no correlation between PANDAR expression and the common gene mutations (Table 1, P>0.05).
Effect of PANDAR expression on chemotherapy response in AML
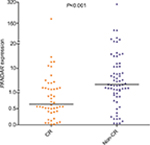
In order to explore the impact of PANDAR expression in clinical prognosis with AML patients, we analyzed 115 AML patients with available follow-up data. Compared with PANDARlow group, patients in PANDARhigh group had a lower CR rate (P<0.001, Table 1). We then analyzed the expression level of PANDAR in AML patients who achieved CR and those without CR, and showed it in scatter plots (P<0.001, Figure 3). Additionally, clinical characteristics of patients with CR and non-CR were further compared. Significant differences were found in PANDAR expression, age, WBCs, BM blast, risk group, and karyotype finding (P<0.05, Table 2). Logistic regression analysis including the most predictive factors was further performed which revealed that PANDAR expression was an independent risk factor that affected CR in whole-cohort AML and non-M3 AML patients (P=0.010 and 0.005, respectively, Tables 3 and 4).
The relationship between PANDAR expression and prognosis in AML patients
The survival analysis indicated that in the whole-cohort AML patients with high PANDAR expression had a shorter overall survival (OS) time than those who were in PANDAR low-expressed group (P<0.001, Figure 4A). In non-M3 AML, patients with PANDAR high expression also had a shorter OS compared with those with PANDAR low expression (P=0.005, Figure 4B). Regretfully, patients with PANDAR high expression did not presented a significant shorter OS than patients with PANDAR low expression among CN-AML (P=0.238, Figure 4C). Multivariate analysis which included variables in univariate analysis with P<0.2 (WBC [≥30×109/L vs <30×109/L], age [≤60 vs >60 years], risk group [favorable vs intermediate vs poor], PANDAR expression [high vs low], gene mutations [mutant vs wild type]). Multivariate analysis further showed that PANDAR expression was a significant independent risk factor in affecting OS among whole-cohort AML patients and non-M3 AML patients (P=0.033 and 0.032, respectively, Tables 3 and 4).
Discussion
Lately, more and more researchers are devoted to exploring noncoding RNA and AML.26 Many studies have proved that lncRNAs indeed played an important regulatory role in human cancers, and it was closely related with the occurrence and the development of various tumors.14,27 Also, increasing number of research papers have shown that the abnormal expression of PANDAR was connected with the tumorigenesis of various solid tumors.28–32 In the first report published by Hung et al, it was indicated that PANDAR inhibited the expression of proapoptotic genes by interacting with the transcription factor NF-YA.12 Thereafter, Li et al28 found that PANDAR was upregulated in thyroid cancer. Further investigating the regulatory mechanism of PANDAR, Li et al28 also found that knockdown of PANDAR could promote apoptosis of thyroid cells by reducing the expression of Bcl2 and activating Bax. In addition, Sang et al30 also reported that PANDAR, which was obviously upregulated in breast cancer tissues and cell lines, could affect the cell cycle by regulating its downstream target p16INK4A. In summary, PANDAR played a significant role in various cancers, including in cancer initiation and progression, and it could serve as an oncogene in these cancers.
In the studies examining the expression level of PANDAR, many reports showed that PANDAR was associated with the prognosis of cancers. For instance, Li et al found that PANDAR was upregulated in thyroid cancer tissue and cell lines, and it could be a promising therapeutic target and important biomarker for thyroid cancer.28 Similarly, an article reported that the expression level of PANDAR in hepatocellular carcinoma was crucially associated with the size of tumor nodule, vascular invasion, and TNM stage.29 Moreover, overexpression of PANDAR was relevant to the poorer survival and shorter recurrence duration for the disease in hepatocellular carcinoma patients, and it could be recognized as a potential tumor biomarker and therapeutic target.29 However, the effect of PANDAR expression on prognosis in blood malignancies remains poorly defined. Findings from our study demonstrated that high expression of PANDAR indicated a poor prognosis in AML patients. PANDAR expression level influenced CR rate, with the PANDARhigh group having lower CR rate in comparison to the PANDARlow group. Logistic regression analysis showed that PANDAR expression was an independent prognostic factor for CR. More importantly, Kaplan–Meier survival analyses clearly showed that patients with higher expression of PANDAR had a shorter OS than those patients with lower expression. Univariate and multivariate Cox regression analyses revealed the increased PANDAR expression was an independent unfavorable risk factor in AML patients.
Our study was the first to report that PANDAR was upregulated in AML and was also the first to demonstrate the prognostic value of PANDAR in AML.
Conclusion
Expression of PANDAR was frequently upregulated in AML, and high expression of PANDAR as an independent unfavorable risk factor for CR and OS in whole-cohort and non-M3 AML patients. Therefore, our findings indicated that PANDAR was a potential biomarker for AML and it might effectively predict the outcome of AML patients.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81270630), Medical Innovation Team of Jiangsu Province (CXTDB2017002), 333 Project of Jiangsu Province (BRA2016131), Six Talent Peaks Project in Jiangsu Province (2015-WSN-115), Zhenjiang Clinical Research Center of Hematology (SS2018009), China Postdoctoral Science Foundation funded project (2016M601748), Youth Medical Talents Project of “Ke Jiao Qiang Wei” project of Jiangsu province (QNRC2016450, QNRC2016449), and Key Medical Talent Program of Zhenjiang City.
Disclosure
The authors report no conflicts of interest in this work.
References
Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373(12):1136–1152. | ||
Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. | ||
Ferrara F. Unanswered questions in acute myeloid leukaemia. Lancet Oncol. 2004;5(7):443–450. | ||
Avivi I, Rowe JM. Prognostic factors in acute myeloid leukemia. Curr Opin Hematol. 2005;12(1):62–67. | ||
Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14(3):497–529. | ||
Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100(13):4325–4336. | ||
Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109(2):431–448. | ||
Halley P, Kadakkuzha BM, Faghihi MA, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6(1):222–230. | ||
Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. | ||
Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–2425. | ||
Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. | ||
Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. | ||
Li J, Li Z, Zheng W, et al. PANDAR: a pivotal cancer-related long non-coding RNA in human cancers. Mol Biosyst. 2017;13(11):2195–2201. | ||
Zhou JD, Zhang TJ, Li XX, et al. Epigenetic dysregulation of ID4 predicts disease progression and treatment outcome in myeloid malignancies. J Cell Mol Med. 2017;21(8):1468–1481. | ||
Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. | ||
Lin J, Yao DM, Qian J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6(10):e26906. | ||
Yang X, Qian J, Sun A, et al. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46(7–8):579–583. | ||
Qian J, Yao DM, Lin J, et al. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7(9):e45760. | ||
Lin J, Yao DM, Qian J, et al. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91(4):519–525. | ||
Lin J, Qian J, Yao DM, et al. Rapid and reliable detection of IDH1 R132 mutations in acute myeloid leukemia using high-resolution melting curve analysis. Clin Biochem. 2011;44(10–11):779–783. | ||
Wen XM, Lin J, Yang J, et al. Double CEBPA mutations are prognostically favorable in non-M3 acute myeloid leukemia patients with wild-type NPM1 and FLT3-ITD. Int J Clin Exp Pathol. 2014;7(10):6832–6840. | ||
Wen XM, Hu JB, Yang J, et al. CEBPA methylation and mutation in myelodysplastic syndrome. Med Oncol. 2015;32(7):192. | ||
Zhou JD, Yang L, Zhang YY, et al. Overexpression of BAALC: clinical significance in Chinese de novo acute myeloid leukemia. Med Oncol. 2015;32(1):386. | ||
Wang YX, Zhang TJ, Yang DQ, et al. Reduced miR-215 expression predicts poor prognosis in patients with acute myeloid leukemia. Jpn J Clin Oncol. 2016;46(4):350–356. | ||
Zhou JD, Lin J, Zhang TJ, et al. Hypomethylation-mediated H19 overexpression increases the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukemia. J Cell Physiol. 2018;233(3):2444–2450. | ||
Zhou JD, Zhang LC, Zhang TJ, et al. Dysregulation of miR-200s clusters as potential prognostic biomarkers in acute myeloid leukemia. J Transl Med. 2018;16(1):135. | ||
Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: there’s a long way to go. J Mammary Gland Biol Neoplasia. 2012;17(1):43–58. | ||
Li Z, Gao B, Hao S, et al. Knockdown of lncRNA-PANDAR suppresses the proliferation, cell cycle and promotes apoptosis in thyroid cancer cells. Excli J. 2017;16:354–362. | ||
Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother. 2015;72:113–118. | ||
Sang Y, Tang J, Li S, et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16INK4A expression. Sci Rep. 2016;6:22366. | ||
Ma P, Xu T, Huang M, Shu Y. Increased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancer. Biomed Pharmacother. 2016;78:172–176. | ||
Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol. 2017;143(1):71–81. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.