Back to Journals » Cancer Management and Research » Volume 12
Overexpression of lncRNA NLIPMT Inhibits Colorectal Cancer Cell Migration and Invasion by Downregulating TGF-β1
Authors An Y, Zhang S, Zhang J, Yin Q, Han H, Wu F, Zhang X
Received 30 January 2020
Accepted for publication 18 June 2020
Published 20 July 2020 Volume 2020:12 Pages 6045—6052
DOI https://doi.org/10.2147/CMAR.S247764
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Yongkang An,1 Shuangxi Zhang,1 Jing Zhang,1 Qing Yin,2 Haitao Han,1 Fang Wu,3 Xiangan Zhang1
1The First Affiliated Hospital of Henan University of TCM, Anorectal Disease Clinic, Zhengzhou City, Henan Province 450000, People’s Republic of China; 2The Affiliated Hospital of Henan Academy of TCM, Nursing Department, Zhengzhou City, Henan Province 450000, People’s Republic of China; 3The First Affiliated Hospital of Henan University of TCM, Medical and Nursing Joint, Zhengzhou City, Henan Province 450000, People’s Republic of China
Correspondence: Xiangan Zhang
The First Affiliated Hospital of Henan University of TCM, Anorectal Disease Clinic, Zhengzhou City, Henan Province 450000, People’s Republic of China
Email [email protected]
Background: NLIPMT, as a tumor suppressive lncRNA, has only been investigated in breast cancer, while its roles in other types of cancer remain unknown. This study aimed to explore the role of NLIPMT in colorectal cancer (CRC).
Methods: Expression levels of NLIPMT and TGF-β 1 in two types of CRC tissue (Non-tumor tissues and tumor tissues) were measured and compared by qRT-PCR and paired t-test, respectively. Correlations between the expression of NLIPMT and TGF-β 1 were analyzed by performing linear regression. The effects of transfections on cell invasion and migration were evaluated by Transwell assays.
Results: We found that NLIPMT was downregulated, while TGF-β 1 was upregulated in CRC. In CRC tumor, a negative correlation was found between the expression of NLIPMT and TGF-β 1. In CRC cells, overexpression of NLIPMT resulted in downregulation, while silencing of NLIPMT resulted in upregulation of TGF-β 1. Analysis of cell invasion and migration showed that overexpression of NLIPMT suppressed the tumor cell invasion and migration. In contrast, overexpression of TGF-β 1 could promote CRC cell invasion and migration and also reduce the role of NLIPMT. Through the overall survival evaluation, NLIPMT-high groups of CRC represented better survival rate compared to that of the NLIPMT-low group patients.
Conclusion: The expression of lncRNA NLIPMT was negatively correlated with TGF-β 1 in CRC. Overexpression of NLIPMT inhibited the colorectal cancer cell migration and invasion by downregulating TGF-β 1. Furthermore, the expression of NLIPMT in CRC patients predicted better prognosis, which suggested that NLIPMT could be considered as a novel diagnosis biomarker.
Keywords: NLIPMT, colorectal cancer, TGF-β 1, invasion, migration
Introduction
According to the latest cancer statistics, colorectal cancer (CRC) could affect more than 140,000 new cases and cause more than 50,000 deaths in 2019.1 Moreover, both incidence and mortality rates of CRC are predicted to be continuously increasing.2 The existence of distant metastasis that is caused by the low early diagnostic rate, and development of drug resistance during chemotherapy, are the two major causes of deaths among CRC patients.3 Therefore, novel molecular markers are needed to improve the diagnosis and treatment of CRC.4 However, molecular pathogenesis of this disease is barely known,5,6 which is the major challenge in development of novel diagnostic and therapeutic markers.
Transforming growth factor (TGF)-β plays pivotal roles in diverse cellular processes, such as cell apoptosis, invasion, differentiation as well as cell growth arrest.7 In cancer biology, TGF-β induces epithelial-mesenchymal transition and promote cancer cell invasion and migration.7,8 Therefore, inhibition of TGF-β is considered as a potential therapeutic option for many types of cancer, such as CRC.9 It is known that TGF-β signaling can be regulated by long non-coding RNAs (lncRNAs, >200 nt),10 which participate in cancer biology mainly by regulating gene expression.11 A recent study reported a novel lncRNA named NLIPMT (also known as RP11-115N4.1) as a tumor suppressor in breast cancer.12 In breast cancer, NLIPMT inactivates glycogen synthase kinase 3β (GSK-3β) to suppress cancer metastasis.12 It is known that GSK-3β can regulate TGF-β.13 Therefore, NLIPMT may also interact with TGF-β. This study aimed to investigate the interaction between TGF-β1 and NLIPMT in CRC.
Materials and Methods
CRC Patients and Tissue Samples
This study selected 66 CRC patients from the 115 CRC cases admitted to the First Affiliated Hospital of Henan University of TCM between June 2012 and May 2014. This study passed the review board of aforementioned hospital. Inclusion criteria were: 1) newly diagnosed CRC cases; 2) no initiated therapies were observed; 3) completed treatment and a 5-year follow-up. Exclusion criteria were: 1) patients diagnosed with multiple clinical disorders; 2) recurrent CRC patients. AJCC system was used to stage all the 66 patients, and the results revealed 9, 12, 23 and 22 cases at stage I–IV, respectively. Before this study, informed consent was signed by all selected patients.
All selected patients were diagnosed by MRI-guided colorectal biopsy (the clinical characteristic data not shown). During biopsy, CRC and non-tumor (within 2 cm around tumors) tissues were obtained from these patients. Based on the results of histopathological examinations, all CRC tissues contained more than 90% cancer cells and all non-tumor tissues contained less than 5% cancer cells.
Follow-Up and Treatment
From the day of admission, all patients were followed-up for 5 years to monitor their survival conditions. Patients were treated with surgical resections and/or chemotherapy, targeted therapy or radiotherapy according to their situations. Patients failed to complete follow-up and patients died of accidents or other diseases were excluded. Based on the median level of NLIPMT in CRC tissues, patients were grouped into high and low (n = 33) level groups.
CRC Cells and Transfection
Human CRC cell lines RKO (ATCC, USA) and HCT-116 (ATCC, USA) were used. A mixture of 90% Eagle’s Minimum Essential Medium (EMEM) (Cat no. 30–2003, ATCC) and 10% fetal bovine serum (FBS, Gibco, USA) was used to cultivate RKO and HCT-116 cells, and the culture conditions were 95% humidity, 37 °C and 5% CO2. Cells were harvested at a confluence of 70–80% to perform subsequent experiments. NLIPMT and TGF-β1 expression vector, as well as negative control (NC) siRNA and NLIPMT siRNA were purchased from GenePharma (Shanghai, China). Lipofectamine 2000 reagent (GenePharma, Shanghai, China) was used to transfect 45 nM siRNA (NC siRNA as NC) or 10 nM vector (empty vector as NC) into 1×106 cells. Untransfected cells were used as control (C) cells for all transfections. Cells harvested at 24 h post-transfection were used to carry out the following experiments.
qRT-PCR
Cells harvested at 24 h post-transfection were counted, and Trizol reagent (Invitrogen, USA) was used to extract total RNAs from 6×105 cells. After digestion by DNase I at 37 °C for 1h, RNA samples were subjected to reverse transcriptions, which were performed using QuantiTect Reverse Transcription Kit (QIAGEN, USA). With GAPDH as the endogenous control, DyNAmo Flash SYBR Green qPCR Kit (Thermo Fisher Scientific) was used to prepare all QRT-PCR mixtures to assay the expression of NLIPMT and TGF-β1 mRNA. The primer sequences were listed as following: NLIPMT-Forward primer: 5ʹ-GCCCACCACAACCATTCTT-3ʹ, NLIPMT-Reverse primer: 5ʹ-CAATGGCTGGAGGAAGAGGA-3ʹ; TGF-β1-Forward primer: 5ʹ-TTGAGACTTTTCCGTTGCCG-3ʹ, TGF-β1-Reverse primer: 5ʹ- CGAGGTCTGGGGAAAAGTCT-3ʹ; GAPDH-Forward primer: 5ʹ- CACATCGCTCAGACACCATG-3ʹ, GAPDH-Reverse primer: 5ʹ- TGACGGTGCCATGGAATTTG-3ʹ; All data were processed by using 2−ΔΔCT method, and each experiment were repeated 3 times. (note: *: p < 0.05)
Western Blot (WB)
Cells harvested at 24 h post-transfection were counted, and RIPA solution (GenePhram, Shanghai, China) was used to extract total proteins from 1×106 cells. Following protein denaturing at 95 °C for 10 min, SDS-PAGE (10%) gels were used to carry out electrophoresis to separate proteins. Proteins were then transferred to PVDF membranes and blocked the non-specific binding in blocking buffer (5% BSA) at room temperature for 2 h. After that, the first incubation was performed using rabbit primary antibodies of anti-TGF-β1 (1:1200, ab92486, Abcam) and anti-GAPDH antibody (1:1200, ab9485, Abcam) at 4 °C for 15 h, followed by the second incubation with secondary antibody of anti-rabbit IgG-HRP (1:1500, MBS435036, MyBioSource) at 24 °C for 2 h. Membranes were incubated with ECL (Sigma-Aldrich) for 10 min to develop signals and gray values were analyzed using Image J V1.34 software.
Cell Invasion and Migration
Cells harvested at 24 h post-transfection were counted, and 3×104 cells were mixed with 1 mL aforementioned cell culture medium (serum-free) to prepare single cell suspensions. Transwell assays were performed to analyze the effects of transfections on cell invasion and wound-healing assays were performed to analyze cell migration. Before invasion assay, Matrigel (356,234, Millipore, USA) was used to block the membranes at 37 °C for 6 h. Briefly, the upper chamber was filled with cell suspension and the lower chamber was filled with cell culture medium containing 20% FBS. Under aforementioned conditions, cells were cultivated for 12 h and 0.5% crystal violet (Sigma-Aldrich) was used to stain membranes for 15 min. Finally, an optical microscope was used to observe the invading cells. Meanwhile, the same cells were inoculated into 6 well plate, when the confluency reached 80%, creating a “wound” scratch with a 20 μL tip on the cell monolayer. After washing for 2 times with warmed PBS, cells were cultured for another 48 h with serum-free medium. Photos were taken with phase contrast microscope (Olympus) to assess the scratch healing difference. All experiments were repeated at least three times.
Statistical Analysis
GraphPad Prism software (version 8.3, GraphPad Inc., San Diego, CA, USA) was used for data analysis. Mean ± standard deviation (SD) represented the data for at least three replicates. Linear regression was used to analyze correlations between different genes. Differences among cell transfection groups were compared by ANOVA (one-way) and Tukey’s test. Differences between non-tumor and CRC tissues were compared by paired t-test. Survival curves were plotted and compared by K-M plotter and Log-rank test, respectively. p < 0.05 was statistically significant.
Results
The Expression of NLIPMT and TGF-β1 Were Altered in CRC
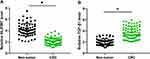
The expression levels of NLIPMT and TGF-β1 in two types of tissue (non-tumor and CRC) were measured and compared by qRT-PCR and paired t-test, respectively. We observed that the expression levels of NLIPMT in CRC tissues were significantly decreased than that in non-tumor tissues (Figure 1A, p < 0.05). In contrast, the expression levels of TGF-β1 were significantly higher in CRC than that in non-tumor tissues (Figure 1B, p < 0.05). These results were consistent with previously studies.8
NLIPMT and TGF-β1 Were Negatively Correlated in CRC Tissues
Correlations between the expression of NLIPMT and TGF-β1 were analyzed by linear regression using GraphPad Prism analysis. It showed that the expression levels of NLIPMT were inversely correlated with that of TGF-β1 in CRC tissues (p < 0.001, Figure 2A). However, NLIPMT and TGF-β1 did not have a significantly correlation in non-tumor tissues (p = 0.4214, Figure 2B). These results suggested that NLIPMT could regulate TGF-β1 directly or indirectly in colorectal cancer cells.
Low Expression Levels of NLIPMT Predicted Poor Survival
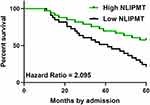
To detect the potential role of NLIPMT in patients survival, we divided the patients into NLIPMT-high and NLIPMT-low groups, then compared the survival curves between these two groups. It showed that the overall survival rate of CRC patients during the 5 years survival was significantly higher in the high NLIPMT level group than that in the low NLIPMT level group (Figure 3, p = 0.03), which suggested that NLIPMT could be considered as a maker for better prognosis.
NLIPMT Downregulated TGF-β1 in CRC Cell Lines
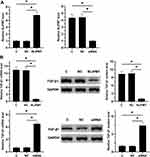
RKO cells were transfected with NLIPMT expression vector and NLIPMT siRNA. Then qRT-PCR was performed to measure the expression levels of NLIPMT at 24 h post-transfection. NLIPMT expression vector could significantly increase the expression levels of NLIPMT and silencing of NLIPMT successfully knocked-down its expression in RKO cells (Figure 4A, p < 0.05). Similar results were also obtained in HCT-116 cells (Supplementary Figure 1A). Moreover, overexpression of NLIPMT inhibited the expression of TGF-β1 in RKO cell lines after transfected with NLIPMT expression vector at both mRNA and protein levels. Meanwhile, silencing of NLIPMT inversely rescued the expression of TGF-β1 at both mRNA and protein levels (Figure 4B, p < 0.05). Similar results were also obtained in HCT-116 cells (Supplementary Figure 1B). All these results indicated that NLIPMT could downregulate TGF-β1 in CRC cells.
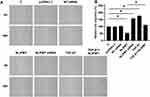
NLIPMT Inhibited Invasion and Migration of Tumor Cells Through TGF-β1
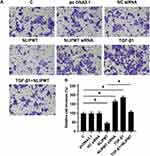
The effects of NLIPMT on cell invasion and migration were evaluated by Transwell assays and wound-healing assays. Compared to C and NC groups, overexpression of NLIPMT suppressed the tumor cells invasion (Figure 5A and B) and migration (Figure 6A and B) in RKO cells, while silencing of NLIPMT played an opposite role (p < 0.05). Moreover, overexpression of TGF-β1 promoted CRC cell invasion (Figure 5A and B) and migration (Figure 6A and B), and also reduced the role of overexpressing NLIPMT (p < 0.05). And similar effects were also observed in HCT-116 cells (data not shown). Therefore, lncRNA NLIPMT might function as a tumor suppressor in CRC.
Discussion
The functionality and prognostic value of NLIPMT in CRC were explored in this paper. We found that NLIPMT was downregulated in CRC and predicted the poor survival of CRC patients. Moreover, NLIPMT may negatively regulate TGF-β1 to suppress the invasion and migration of CRC cells.
At present, the prediction of the survival of cancer patients is mainly based on clinical stages, which were determined by tumor size, lymph node metastasis, and distant metastasis.14 Development of novel prognostic factors may guide the selection of treatment options and the prediction of outcomes, which in turn benefit patients’ survival.14 The expression pattern and functionality of NLIPMT have only been investigated in breast cancer, in which NLIPMT was downregulated, and overexpression of NLIPMT was shown to inhibit cancer metastasis.12 This study is the first to report the downregulation of NLIPMT in CRC and the increased invasion and migration rates of CRC cells. Therefore, NLIPMT is also a tumor suppressive lncRNA in CRC. In addition, low expression levels of NLIPMT in CRC tissues were proved to be closely correlated with the poor prognosis of CRC patients. Therefore, measurement of the expression levels of NLIPMT before therapies may guide the prognostic assignment of CRC patients. However, the accuracy remains to be further verified.
TGF-β1 is overexpressed in CRC to induce cancer metastasis.15,16 Therefore, multiple anti-cancer drugs have been developed to target TGF-β1.15,16 Consistently, this study also observed the overexpression of TGF-β1 in CRC tissues. In addition, overexpression of TGF-β1 resulted in increased invasion and migration rates of CRC cells. Interestingly, we showed that NLIPMT negatively correlated with TGF-β1 in CRC cells. The possible explanation is that NLIPMT can inactive GSK-3β,12 while GSK-3β inactivation can lead to the downregulation of TGF-β1.13 It is known that GSK-3β is activated in CRC.17 Therefore, activation of GSK-3β in CRC may mediate the interaction between NLIPMT and TGF-β1. This is also one possible explanation of the lacking of significant correlation between NLIPMT and TGF-β1 in non-tumor tissues. Moreover, lncRNA could regulate the protein-coding RNA in many ways, some other transcription factors or enzymes might be involved in the NLIPMT- TGF-β1 signaling. Therefore, more evidence is needed to clarify the complicated regulation mechanism between NLIPMT and TGF-β1.
Conclusions
In conclusion, NLIPMT is overexpressed in CRC and negatively regulates TGF-β1 to suppress CRC cell invasion and migration. NLIPMT could be considered as a potential prognostic biomarker for the diagnosis and treatment of CRC patients.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Henan University of TCM. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent prior to their inclusion within the study.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. DeSantis CE, Miller KD, Goding Sauer A, et al. Cancer statistics for African Americans, 2019. CA Cancer J Clin. 2019;69(3):211–233. doi:10.3322/caac.21555
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Sasaki H, Miura K, Horii A, et al. Orthotopic implantation mouse model and cDNA microarray analysis indicates several genes potentially involved in lymph node metastasis of colorectal cancer. Cancer Sci. 2008;99(4):711–719. doi:10.1111/j.1349-7006.2008.00725.x
4. Gupta RA, DuBois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1(1):11–21. doi:10.1038/35094017
5. Yamagishi H, Kuroda H, Imai Y, et al. Molecular pathogenesis of sporadic colorectal cancers. Chin J Cancer. 2016;35(1):4. doi:10.1186/s40880-015-0066-y
6. Dahmus JD, Kotler DL, Kastenberg DM, et al. The gut microbiome and colorectal cancer: a review of bacterial pathogenesis. J Gastrointest Oncol. 2018;9(4):769–777. doi:10.21037/jgo.2018.04.07
7. Miyazono K, Katsuno Y, Koinuma D, et al. Intracellular and extracellular TGF-β signaling in cancer: some recent topics. Front Med. 2018;12(4):387–411. doi:10.1007/s11684-018-0646-8
8. Visan I. Targeting TGF-β in cancer. Nat Immunol. 2018;19(4):316.
9. Tiwari A, Saraf S, Verma A, et al. Novel targeting approaches and signaling pathways of colorectal cancer: an insight. World J Gastroenterol. 2018;24(39):4428–4435. doi:10.3748/wjg.v24.i39.4428
10. Wang X, Lai Q, He J, et al. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16(1):51–59. doi:10.7150/ijms.27359
11. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
12. Jiang Y, Lin L, Zhong S, et al. Overexpression of novel lncRNA NLIPMT inhibits metastasis by reducing phosphorylated glycogen synthase kinase 3β in breast cancer. J Cell Physiol. 2019;234(7):10698–10708. doi:10.1002/jcp.27738
13. Potz BA, Scrimgeour LA, Sabe SA, et al. Glycogen synthase kinase 3β inhibition reduces mitochondrial oxidative stress in chronic myocardial ischemia. J Thorac Cardiovasc Surg. 2018;155(6):2492–2503. doi:10.1016/j.jtcvs.2017.12.127
14. Hueman MT, Wang H, Yang CQ, et al. Creating prognostic systems for cancer patients: a demonstration using breast cancer. Cancer Med. 2018;7(8):3611–3621. doi:10.1002/cam4.1629
15. Han S, Bui NT, Ho MT, et al. Dexamethasone inhibits TGF-β1–induced cell migration by regulating the ERK and AKT pathways in human colon cancer cells via CYR61. Cancer Res Treat. 2016;48(3):1141–1153. doi:10.4143/crt.2015.209
16. Dai G, Sun B, Gong T, et al. Ginsenoside Rb2 inhibits epithelial-mesenchymal transition of colorectal cancer cells by suppressing TGF-β/Smad signaling. Phytomedicine. 2019;56:126–135. doi:10.1016/j.phymed.2018.10.025
17. Chen JS, Huang JQ, Luo B, et al. PIK3CD induces cell growth and invasion by activating AKT/GSK‐3β/β‐catenin signaling in colorectal cancer. Cancer Sci. 2019;110(3):997–1011. doi:10.1111/cas.13931
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.