Back to Journals » OncoTargets and Therapy » Volume 11
Overexpression of HDAC9 is associated with poor prognosis and tumor progression of breast cancer in Chinese females
Authors Huang Y, Jian W, Zhao J, Wang G
Received 4 February 2018
Accepted for publication 1 March 2018
Published 17 April 2018 Volume 2018:11 Pages 2177—2184
DOI https://doi.org/10.2147/OTT.S164583
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Yixiang Huang,* Wei Jian,* Junyong Zhao, Gang Wang
Department of General Surgery, Tenth People’s Hospital of Tongji University, Shanghai, China
*These authors contributed equally to this work
Background: Breast cancer represents a serious health issue among females. HDAC9 has been identified as an oncogene in human cancers. This study sought to assess the prognostic value and the biologic function of HDAC9 in breast cancer patients.
Methods: Expression of HDAC9 in breast cancer tissues and cells was evaluated by quantitative real-time polymerase chain reaction. Kaplan–Meier survival analysis and Cox regression assay were conducted to explore the prognostic significance of HDAC9. Cell experiments were performed to investigate the effects of HDAC9 on the biologic behaviors of breast cancer cells.
Results: Expression of HDAC9 was significantly upregulated in both cancerous tissues and cells compared with the normal controls (all P<0.05). Overexpression of HDAC9 was correlated with lymph node metastasis (P=0.021) and TNM stage (P=0.004). Patients with high HDAC9 had poor overall survival compared to those with low levels of HDAC9 (log-rank P<0.05). Elevated HDAC9 was found to be an independent prognostic factor for the patients (hazard ratio=2.996, 95% CI=1.611–5.572, P=0.001). According to the cell experiments, tumor cell proliferation, migration and invasion were suppressed by knockdown of HDAC9.
Conclusion: All data demonstrated that overexpression of HDAC9 serves as a prognostic biomarker and may be involved in the tumor progression of breast cancer.
Keywords: HDAC9, prognosis, progression, breast cancer
Introduction
Breast cancer is the most common malignancy among females around the world.1 Patients suffering from breast cancer usually have the symptoms of change in breast shape, fluid coming from the nipple, dimpling of skin or a red scaly patch of skin.2,3 Every year, ~1,300,000 cases are diagnosed with breast cancer and about 465,000 deaths are estimated to occur worldwide.4 Some risk factors have been identified to be correlated with the occurrence of breast cancer, including obesity, drinking alcohol, lack of physical exercise, ionizing radiation, early age at first menstruation, family history and older age.5 Despite advances in surgery, chemotherapy and radiotherapy, the prognosis of breast cancer remains dismal.6 Data from previous studies revealed that the prognosis could be achieved by using some molecular biomarkers.7 It is, therefore, necessary to uncover more precise prognostic biomarkers for determining prognosis in patients with breast cancer.
It is generally considered that histone acetyltransferase and histone deacetylase (HDAC) play important roles during the regulation of gene transcription.8,9 Currently, the relationship between HDACs and progression of cancer has attracted attention in different malignancies. Data in several previous studies reveal that an aberrant expression of HDACs has been detected in many tumor samples.10,11 In addition, HDACs have been found to inhibit tumor suppressor expression by binding to the promoter region.12,13 HDAC9, a subtype of HDAC, has been investigated in some types of human cancers. For example, Moreno et al demonstrated that the expression of HDAC9 was deregulated and it correlated with overall survival of patients with lymphoblastic leukemia.14 However, reports about the role of HDAC9 in breast cancer are currently limited.
To better understand the relationship of HDAC9 and breast cancer, this study examined the expression patterns and prognostic significance of HDAC9 in breast cancer patients. The effects of HDAC9 on biologic behaviors of cancer cells were also assessed.
Materials and methods
Patients and tissue sample collection
Tissue specimens used for the subsequent experiments were collected from 118 breast cancer patients who underwent surgery between 2007 and 2011 at the hospital and were verified by experienced pathologists. None of these patients had received any antitumor therapy before the sampling. The breast cancer tissues and the adjacent normal tissues were snap-frozen in liquid nitrogen after collection. Signed informed consent was obtained from each patient, and this study was approved by the Ethics Committee of Tenth People’s Hospital of Tongji University. Moreover, the clinicopathologic information on age, tumor size, estrogen receptor status, progesterone receptor status, human epidermal growth factor receptor 2 status, lymph node metastasis and TNM stage was recorded from the electronic medical records of the patients and are summarized in Table 1. After surgery, all the patients were enrolled in a 5-year follow-up survey. The survival information was obtained for the subsequent survival analysis.
Cell lines and transfection
Human breast cancer cell lines MCF-7, BT474 and normal human breast epithelial cell line MCF-10A were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All these cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% fetal bovine serum and kept in a humidified incubator with 5% CO2 at cell transfection was conducted by using Lipofectamine 2000 Reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the instructions of the manufacturer. Cancer cells transfected with HDAC9 siRNA were defined as the experimental group and the other cells treated with control siRNA or Lipofectamine 2000 were used as the control group.
HDAC9 inhibition
In addition to siRNA, vorinostat (suberoylanilide hydroxamic acid [SAHA]) was also used to suppress the expression of HDAC9 in breast cancer cells. MCF-7 and BT474 were seeded in a 96-well culture plate and treated with 10 μM SAHA (vorinostat; MedChem Express Co., Monmouth Junction, NJ, USA) or vehicle (dimethyl sulfoxide [DMSO], 1:1,000) for 48 h. The drug and the medium were replenished every 24 h.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the tissues and cells using TRIzol reagent (Thermo Fisher Scientific) as per the manufacturer’s instructions. Pure RNA was obtained by calculating the ratio of OD A260/280, which was used to indicate the purity of RNA. Reverse transcription was performed to synthesize cDNA from RNA by Transcriptor First Strand cDNA Synthesis Kit (Roche, Vilvoorde, Belgium). Expression of HDAC9 was investigated by using qRT-PCR, which was carried out with the SYBR green I Master Mix kit (Thermo Fisher Scientific) and was run on the 7300 Real-Time PCR System (Thermo Fisher Scientific). The primer sequences of HDAC9 were as follows: forward: 5′-AACTGGAGCAGCAGAGGCAAG-3′, reverse: 5′-TACTTCTGTACTTGCCACTGCC-3′. Besides, GAPDH was used as the internal control with the following primers: forward: 5′-GGCCTCCAAGGAGTAAGACC-3′, reverse: 5′-AGGGGTCTACATGGCAACTG-3′. The final relative expression of HDAC9 was calculated with 2−ΔΔCt method and normalized to GAPDH.
Cell proliferation assay
In order to examine the effects of HDAC9 on cell proliferation of breast cancer cells, the colorimetric MTT analysis was carried out in this study. MCF-7 and BT474 cells were seeded in two 96-well culture plates. Cells in one plate were transfected with HDAC9 siRNA or control vectors (control siRNA or Lipofectamine 2000). Meanwhile, the cells in another plate were treated with SAHA or DMSO for 48 h. After the treatment, each well was added with 10 μL MTT (5 mg/mL; Sigma-Aldrich) and incubated at 37°C for 4 h. Then, 100 μL DMSO (Sigma-Aldrich) was added to the wells to dissolve the formazan crystals. The absorbance value was measured at 490 nm with a spectrophotometer (Multiskan MK3; Thermo Fisher Scientific). Experiments were repeated in triplicate.
Cell migration and invasion analysis
To uncover the effects of HDAC9 on cell migration and invasion, the Transwell analysis was carried out with a 24-well Transwell chamber. The cells transfected with siRNA or treated with SAHA were added in the upper compartment with a concentration of 1×105 per well and then incubated in serum-free RPMI-1640 medium at 37°C for 24 h. The lower compartment contained 300 μL RPMI-1640 medium supplemented with 20% fetal bovine serum, which was used as the chemotactic factor. After incubation for 24 h, the cells that migrated to the lower compartment were stained with 0.1% crystal violet and counted by a microscope. For the invasion assay, the upper chamber was coated with Matrigel (BD, Bedford, MA, USA).
Statistical analysis
All the statistical analyses were conducted by SPSS software (SPSS Inc., Chicago, IL, USA), and the data used were expressed as mean±SD. Differences between the two groups were examined by Student’s t-test. Association of HDAC9 with clinicopathologic features was assessed with chi-square test. Survival analysis was conducted using Kaplan–Meier method and log-rank test. Cox regression analysis was adopted to confirm the prognostic performance of HDAC9 for breast cancer patients. Statistical differences with P<0.05 were considered as statistically significant.
Results
HDAC9 expression in tissue specimens and cells
In this study, the expression of HDAC9 in the tissues and cell lines was estimated by qRT-PCR. The analysis results showed that HDAC9 expression was significantly higher in breast cancer tissues than that in the paired normal tissues (P<0.001, Figure 1A). To confirm this result, HDAC9 expression was also evaluated in breast cancer cells, and it was found that its expression was upregulated in breast cancer cells compared with the normal cells (all P<0.05, Figure 1B), which was in accordance with the results in tissue samples.
Association of HDAC9 with the clinical characteristics of breast cancer patients
Chi-square test was used to estimate the relationship between HDAC9 expression and cancer patients’ clinicopathologic data. To facilitate this assay, a cutoff value of mean HDAC9 expression was chosen to be used to classify the patients into low HDAC9 expression group (n=54) and high HDAC9 expression group (n=64). All the analysis results are detailed in Table 1 and reveal that the expression of HDAC9 was influenced by lymph node metastasis (P=0.021) and TNM stage (P=0.004). However, no relationship was found between HDAC9 expression and age, tumor size, estrogen receptor status, progesterone receptor status or human epidermal growth factor receptor 2 status (all P>0.05).
Prognostic significance of HDAC9 for breast cancer
In this study, the relationship between HDAC9 and overall survival of breast cancer patients was also investigated. Based on the survival information obtained from the 5-year follow-up survey, the Kaplan–Meier survival analysis was performed for breast cancer patients. Survival curves shown in Figure 2 reveal that patients with high HDAC9 expression levels had shorter survival time compared to those with low HDAC9 levels (log-rank P<0.05). Furthermore, Cox regression analysis was carried out to examine the factors that might influence the overall survival. The univariate and multivariate Cox analyses results detailed in Table 2 demonstrate that upregulated HDAC9 was closely correlated with poor overall survival and could be used as an independent prognostic factor for patients with breast cancer (hazard ratio=2.996, 95% CI=1.611–5.572, P=0.001).
HDAC9 reduction inhibits proliferation, migration and invasion of breast cancer cells
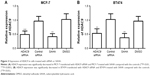
To investigate the functional role of HDAC9 in PCa, its effects on tumor cell proliferation, migration and invasion were examined in this study. Two breast cancer cell lines MCF-7 and BT474 were transfected with HDAC9 siRNA or treated with SAHA to suppress the expression of HDAC9. The results of qRT-PCR showed that the expression of HDAC9 in breast cancer cells transfected with HDAC9 siRNA was significantly lower than that in the cells with control vectors (P<0.01, Figure 3). Similarly, markedly reduced HDAC9 was also detected in the cells treated with SAHA compared to that in those with DMSO (P<0.01, Figure 3). These data indicate that the HDAC9 expression was successfully reduced by siRNA and SAHA. MTT assay was adopted to analyze the cell proliferation, which showed that breast cancer cell proliferation was suppressed in HDAC9-knockdown cells compared with the control cells (P<0.05, Figure 4A and B). In addition to proliferation, cell migration and invasion were also assessed using Transwell analysis. According to the analysis results, the tumor cell migration and invasion were both found to be inhibited in the cells with reduction of HDAC9 compared with the controls (P<0.05, Figure 4C–F).
Discussion
As the most prevalent cancer occurring in women, breast cancer has received lots of attention on its progression and treatment.15 It can be diagnosed at all age groups and has been found to be a great threat to healthy life.16 So far, great progress has been made in therapeutic methods, and the mortality of breast cancer has been reduced.17 However, the prognosis of some cancer cases is not satisfactory mainly due to the advanced stage of the tumors.18 Therefore, prognosis is urgently needed to be improved for breast cancer patients. Data from recent studies show that cancer prognosis has improved due to using related biomarkers, which play crucial roles during tumor progression.19,20 In breast cancer, some prognostic biomarkers have also been identified. For instance, Fu et al demonstrated that SOX17 expression was downregulated and correlated with the poor prognosis of breast cancer.21 MicroRNA-106b, another example, has been proved to be involved in the recurrence of breast cancer and to act as an independent prognostic factor in patients with breast cancer.22 Jerzak et al reported that the thyroid hormone receptor α (THRα) represented an efficient prognostic biomarker in breast cancer patients.23 All these data indicated the pivotal role of cancer-related molecules for breast cancer prognosis. Consequently, more molecular biomarkers should be identified for prediction of prognosis in breast cancer.
During the development of breast cancer, numerous epigenetic changes take place, such as methylation of DNA and diverse histone modifications, including phosphorylation, methylation, sumoylation, ubiquitination and acetylation.24 HDACs represent a series of proteins which play pivotal roles in acetylation.25 Eighteen members of the HDAC family have been identified in human beings. Recent studies have demonstrated that HDACs can be used as therapeutic targets and are involved in tumor progression in different cancers, including breast cancer.26 The altered expression of HDAC1, HDAC5, HDAC6 and HDAC8 has been identified as a reliable prognostic biomarker for breast cancer patients.27–30 HDAC2 and HDAC3 were found to be associated with the aggressive behavior of breast cancer.31 HDAC4 has been reported to mediate the antitumor effects of microRNA-125a-5p in breast cancer.32 HDAC9, also a member of HDAC family, has been found to be deregulated in some tumor samples, such as medulloblastomas and lung cancer.33,34 Moreover, Lapierre et al focused on the functional role of HDAC9 in proliferation of breast cancer cells and found that overexpression of HDAC9 could promote cell proliferation.35 However, the clinical significance of HDAC9 has been rarely reported in patients with breast cancer. To better understand the role of HDAC9, its expression patterns and prognostic significance were assessed in patients with breast cancer.
In this study, the expression of HDAC9 was measured in the breast cancer tissues and cells using qRT-PCR. The analysis results showed that HDAC9 expression was remarkably upregulated in breast cancer tissues and cell lines compared with the normal controls. Moreover, the overexpression of HDAC9 was found to be correlated with lymph node metastasis and TNM stage. Thus, we considered that HDAC9 might act as an oncogene in breast cancer and is involved in the development of this malignancy. Additionally, the clinical significance of HDAC9 in breast cancer prognosis was also investigated. From the Kaplan–Meier survival curves, we found that patients with high HDAC9 expression levels had shorter survival time than those with low HDAC9 levels, suggesting the increased HDAC9 was correlated with poor overall survival. Data in the Cox regression analysis revealed that the upregulated HDAC9 was an independent prognostic factor for patients with breast cancer.
To further confirm the functional role of HDAC9 during breast cancer progression, its effects on biologic behaviors of breast cancer cells were also examined in the current study. In the breast cancer cells, siRNA and SAHA were separately used to silence the expression of HADC9. The analysis results demonstrated that cell proliferation, migration and invasion were suppressed by the knockdown of HDAC9. All the above data suggested that HDAC9 reduction could inhibit tumor progression in breast cancer. In a previous study performed by Lapierre et al,35 overexpression of HDAC9 was reported to promote cell proliferation of breast cancer cells, and it might exert its antitumor effects by targeting sex-determining region Y-box 9 protein (SOX9), which has been described as a crucial molecule during tumor progression of breast cancer by regulating Wnt/β-catenin pathway.36 However, the precise molecular mechanisms underlying the role of HDAC9 in breast cancer need to be confirmed in further studies. The results of this study might be limited by the small sample size, and therefore, further studies are necessary with larger research cohort.
Conclusion
In summary, all data in this study revealed that overexpression of HDAC9 was correlated with the progression of breast cancer and it could be used as a candidate prognostic biomarker.
Disclosure
The authors report no conflicts of interest in this work.
References
DeSantis C, Ma J, Bryan L, et al. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. | ||
Liao MN, Chen SC, Chen SC, et al. Change and predictors of symptom distress in breast cancer patients following the first 4 months after diagnosis. J Formos Med Assoc. 2015;114(3):246–253. | ||
Seber S, Solmaz D, Yetisyigit T. Antihormonal treatment associated musculoskeletal pain in women with breast cancer in the adjuvant setting. Onco Targets Ther. 2016;9:4929–4935. | ||
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–2150. | ||
Ruszczyk M, Zirpoli G, Kumar S, et al. Breast cancer risk factor associations differ for pure versus invasive carcinoma with an in situ component in case-control and case-case analyses. Cancer Causes Control. 2016;27(2):183–198. | ||
Jueckstock J, Kasch F, Jaeger B, et al. Adjuvant therapeutic decisions in elderly breast cancer patients: the role of chemotherapy in a retrospective analysis. Arch Gynecol Obstet. 2015;292(5):1101–1107. | ||
Sun L, Jiang R, Li J, et al. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem. 2017;54(1):127–133. | ||
West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124(1):30–39. | ||
Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12(12):917–930. | ||
Gong F, Miller KM. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res. 2013;750(1–2):23–30. | ||
Sangshetti JN, Sakle NS, Dehghan MH, et al. Histone deacetylases as targets for multiple diseases. Mini Rev Med Chem. 2013;13(7):1005–1026. | ||
Hoang JJ, Baron S, Volle DH, et al. Lipids, LXRs and prostate cancer: are HDACs a new link? Biochem Pharmacol. 2013;86(1):168–174. | ||
Matsuda KI, Mori H, Kawata M. Epigenetic mechanisms are involved in sexual differentiation of the brain. Rev Endocr Metab Disord. 2012;13(3):163–171. | ||
Moreno DA, Scrideli CA, Cortez MA, et al. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;150(6):665–673. | ||
Cuzick J, Sestak I, Thorat MA. Impact of preventive therapy on the risk of breast cancer among women with benign breast disease. Breast. 2015;24 Suppl 2:S51–S55. | ||
Rahimzadeh M, Baghestani AR, Gohari MR, et al. Estimation of the cure rate in Iranian breast cancer patients. Asian Pac J Cancer Prev. 2014;15(12):4839–4842. | ||
Seneviratne S, Campbell I, Scott N, et al. Ethnic differences in timely adjuvant chemotherapy and radiation therapy for breast cancer in New Zealand: a cohort study. BMC Cancer. 2014;14:839. | ||
Chen S, Ibrahim NK, Yan Y, et al. Risk stratification in patients with advanced-stage breast cancer by pretreatment [(18) F]FDG PET/CT. Cancer. 2015;121(22):3965–3974. | ||
Yue X, Lan F, Hu M, et al. Downregulation of serum microRNA-205 as a potential diagnostic and prognostic biomarker for human glioma. J Neurosurg. 2016;124(1):122–128. | ||
Spaks A, Svirina D, Spaka I, et al. CXC chemokine ligand 4 (CXCL4) is predictor of tumour angiogenic activity and prognostic biomarker in non-small cell lung cancer (NSCLC) patients undergoing surgical treatment. Biomarkers. 2016;21(5):474–478. | ||
Fu DY, Tan HS, Wei JL, et al. Decreased expression of SOX17 is associated with tumor progression and poor prognosis in breast cancer. Tumour Biol. 2015;36(10):8025–8034. | ||
Zheng R, Pan L, Gao J, et al. Prognostic value of miR-106b expression in breast cancer patients. J Surg Res. 2015;195(1):158–165. | ||
Jerzak KJ, Cockburn J, Pond GR, et al. Thyroid hormone receptor alpha in breast cancer: prognostic and therapeutic implications. Breast Cancer Res Treat. 2015;149(1):293–301. | ||
Jovanovic J, Ronneberg JA, Tost J, et al. The epigenetics of breast cancer. Mol Oncol. 2010;4(3):242–254. | ||
Ooi JY, Tuano NK, Rafehi H, et al. HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes. Epigenetics. 2015;10(5):418–430. | ||
Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. | ||
Eom M, Oh SS, Lkhagvadorj S, et al. HDAC1 expression in invasive ductal carcinoma of the breast and its value as a good prognostic factor. Korean J Pathol. 2012;46(4):311–317. | ||
Li A, Liu Z, Li M, et al. HDAC5, a potential therapeutic target and prognostic biomarker, promotes proliferation, invasion and migration in human breast cancer. Oncotarget. 2016;7(25):37966–37978. | ||
Zhang Z, Yamashita H, Toyama T, et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res. 2004;10(20):6962–6968. | ||
Hsieh CL, Ma HP, Su CM, et al. Alterations in histone deacetylase 8 lead to cell migration and poor prognosis in breast cancer. Life Sci. 2016;151:7–14. | ||
Muller BM, Jana L, Kasajima A, et al. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer – overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. | ||
Hsieh TH, Hsu CY, Tsai CF, et al. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget. 2015;6(1):494–509. | ||
Milde T, Oehme I, Korshunov A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16(12):3240–3252. | ||
Okudela K, Mitsui H, Suzuki T, et al. Expression of HDAC9 in lung cancer – potential role in lung carcinogenesis. Int J Clin Exp Pathol. 2014;7(1):213–220. | ||
Lapierre M, Linares A, Dalvai M, et al. Histone deacetylase 9 regulates breast cancer cell proliferation and the response to histone deacetylase inhibitors. Oncotarget. 2016;7(15):19693–19708. | ||
Wang H, He L, Ma F, et al. SOX9 regulates low density lipoprotein receptor-related protein 6 (LRP6) and T-cell factor 4 (TCF4) expression and Wnt/beta-catenin activation in breast cancer. J Biol Chem. 2013;288(9):6478–6487. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.