Back to Journals » OncoTargets and Therapy » Volume 9
Overexpression of eIF4E in colorectal cancer patients is associated with liver metastasis
Authors Xu T, Zong Y, Peng L, Kong S, Zhou M, Zou J, Liu J, Miao R, Sun X, Li L
Received 15 October 2015
Accepted for publication 8 December 2015
Published 19 February 2016 Volume 2016:9 Pages 815—822
DOI https://doi.org/10.2147/OTT.S98330
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Daniele Santini
Tao Xu,1 Yuanyuan Zong,2 Lipan Peng,1 Shuai Kong,1 Mingliang Zhou,1 Jianqiang Zou,1 Jinglei Liu,1 Ruizheng Miao,1 Xichao Sun,2 Leping Li1
1Department of Gastrointestinal Surgery, 2Department of Pathology, Shandong Provincial Hospital Affiliated to Shandong University, Jinan, Shandong, People’s Republic of China
Purpose: Liver metastasis is one of the leading causes of death in colorectal cancer (CRC) patients. The present study aimed to evaluate the value of eIF4E as a prognostic marker of colorectal liver metastasis (CLM) and identify the functional role of eIF4E in CRC metastasis.
Patients and methods: The expression level of eIF4E in CRC tissues was analyzed by immunohistochemical staining and Western blot. Expression of eIF4E in CRC cell lines was evaluated by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Western blot. Cell Counting Kit-8 (CCK-8) and Transwell assays were performed to assess the effects of eIF4E on cell proliferation, migration, and invasion. Western blot was further used to investigate the mechanism of eIF4E in tumor metastasis.
Results: The upregulation frequency of eIF4E in the CLM group (82.5%) was higher than that in the non-CLM group (65.0%). Of the 80 patients recruited for the follow-up study, 23 were in the low eIF4E group (ratio of tumor to nontumor tissue < twofold), and 57 were in the high eIF4E group (ratio of tumor to nontumor tissue ≥twofold). In addition, the group exhibiting high eIF4E expression had a higher rate of liver metastasis (47.4%) than the group exhibiting low eIF4E expression (13.0%). In CRC cell lines, the expression of eIF4E was higher than in the normal cells. In vitro functional studies indicated that eIF4E knockdown inhibited the proliferation, migration, and invasion of Lovo and SW480 cells, and suppressed the expression of cyclin D1, VEGF, MMP-2, and MMP-9.
Conclusion: The results of the present study indicated that high eIF4E levels in CRC patients predicted a high risk of liver metastasis. Knockdown of eIF4E inhibited CRC cell metastasis in part through regulating the expression of cyclin D1, VEGF, MMP-2, and MMP-9.
Keywords: eIF4E, colorectal cancer, liver metastasis, functional study
Introduction
Colorectal cancer is the fifth most frequently diagnosed cancer and fourth leading cause of cancer-related mortality in People’s Republic of China.1 According to the data from GLOBOCAN 2012, 253,427 new cases of CRC were diagnosed and 139,416 cases died.2 With the development of new detection methods and treatment strategies, the 5-year survival rate of patients diagnosed with CRC at an early stage has improved to ~80%–90%.3 However, the survival rate decreased to less than 10% if the patient is diagnosed with distant metastases.4 Moreover, ~50% of the CRC patients eventually develop distant metastasis,5 and liver is the most common site of metastatic spread.6 Without treatment, the majority of the CLM patients have a median survival of ~8 months.7 Thus, assessment of markers predicting CLM and insight into the underlying molecular mechanisms of colorectal metastasis are necessary.
eIF4E is a critical component of the eIF4F trimeric translation initiation complex, it regulates the translation of eukaryotic mRNAs by binding to the 5′ cap.8 Because eIF4E is the least abundant component in the complex, its expression level is believed to be rate-limiting for the translation of cap-dependent mRNAs.9 eIF4E is highly expressed in many cancers,10 and overexpression of eIF4E increases cancer susceptibility by preferentially enhancing the translation of a subset of mRNAs implicated in oncogenesis, such as cell proliferation, invasion, angiogenesis, and metastasis.11 Previous studies have identified that cyclin D1, VEGF, and MMPs are involved in the development of cancer metastasis.12,13 Nevertheless, it remains unknown whether eIF4E regulates the translation of cyclin D1, VEGF, and MMPs in CRC progression and whether eIF4E contributes to CLM.
In the current study, we found the level of eIF4E in CLM tissues was much higher than in non-CLM tissues, and patients with high eIF4E expression had a higher risk of liver metastasis. In addition, eIF4E participated in the proliferation, migration, and invasion of CRC cells and regulated the expression of cyclin D1, VEGF, MMP-2, and MMP-9.
Materials and methods
Cell lines, tissue specimen, and patients
CRC cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China), and cultured as described previously.14 Forty pairs of CLM tissue, 40 pairs of non-CLM tissue, and patients recruited for the follow-up assay were obtained from the Department of Gastrointestinal Surgery in Shandong provincial hospital. There were no significant differences in the age, sex, nutritional status, and accompanying diseases between the groups. None of the patients had received preoperative radiation/chemotherapy. All the patients received standard D2 surgical resection, and the tissues were frozen in liquid nitrogen after removal. The end points of the follow-up were the occurrence of CLM and cancer-related death. The study was approved by the Ethical Committee of Shandong Provincial Hospital. Written informed consent was obtained from the participants.
Immunohistochemical analysis
Paraffin sections (5 mm) of the tumor tissue were deparaffinized and hydrated, and then incubated overnight at 4°C with eIF4E rabbit monoclonal antibody (#2067, 1:800, Cell Signaling Technology, Inc., Beverly, MA, USA). Secondary staining with biotin-conjugated anti-rabbit IgG, and tertiary staining with horseradish peroxidase (HRP)-conjugated streptavidin were performed using an ABC kit (PK6100, Vector, Burlingame, CA, USA). The slides were developed with 3,3′-diaminobenzidine and counterstained with hematoxylin. Images were captured using a Nikon ECLIPSE TE2000-S microscope system (Nikon, Melville, NY, USA).
RT-qPCR
Total RNA was extracted with the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, UAS), and reverse-transcribed using a PrimeScript™ RT reagent kit (Takara, Shiga, Japan). Reverse transcription quantitative polymerase chain reaction (qRT-PCR) was performed using a SYBR Premix Ex Taq kit (Takara) on an ABI 7300 (Thermo Fisher Scientific). β-actin was amplified as an endogenous control. The relative mRNA expression level was determined using the 2−ΔΔCt method.15 The primers used were as follows: eIF4E: forward 5′-GAATCTAATCAGGAGGTTGCT-3′, reverse 5′-CAATCACTATCTTTGGAGGAA-3′; β-actin: forward 5′-AAAGACCTGTACGCCAACAC-3′, reverse 5′-GTCATACTCCTGCTTGCTGAT-3′. The PCR cycle conditions were 95°C for 30 seconds, followed by 40 cycles at 95°C for 30 seconds, 56°C for 30 seconds, and 72°C for 30 seconds.
Western blot analysis
The harvested cells and tissues were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Institute of Biotechnology, Shanghai, People’s Republic of China). The total protein levels were quantified with a BCA Protein Assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). After 1 hour of blocking in 5% nonfat milk, the membranes were washed with Tris-buffered saline with Tween 20 (TBST) and incubated overnight at 4°C with the following primary antibodies: rabbit anti-eIF4E (#2067, 1:1,000, Cell Signaling Technology), cyclin D1 (#2922, 1:1,000, Cell Signaling Technology), VEGF (sc-152, Santa Cruz Biotechnology Inc., Dallas, TX, USA), rabbit anti-MMP-2 (#13132, 1:1,000, Cell Signaling Technology), rabbit anti-MMP-9 (#13667, 1:1,000, Cell Signaling Technology), and mouse anti-β-actin (sc-47778, Santa Cruz Biotechnology Inc.). After washing with TBST, the membranes were incubated with goat anti-rabbit (BA1054, 1:5,000, Wuhan Boster Biological Technology, Ltd., Wuhan, People’s Republic of China) and goat anti-mouse (BA1051, 1:5,000, Wuhan Boster Biological Technology, Ltd.) IgG. The protein signals were detected using an enhanced chemiluminescence (ECL) kit (Pierce, Rockford, IL, USA) with exposure to X-ray films (Eastman Kodak, Rochester, NY, USA).
SiRNA transfection
eIF4E and control siRNAs were purchased from Ribobio (Cat No Q000001977-1-B, Guangzhou, People’s Republic of China). Lovo and SW480 cell lines were transfected with 100 nM siRNA or negative control using Lipofectamine 2000 reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The silencing of eIF4E gene was tested by RT-qPCR 48 hours post-transfection, and by Western blot 72 hours post-transfection.
Cell proliferation assay
Cell proliferation was measured using a Cell Counting Kit-8 (CCK-8, Beyotime Institute of Biotechnology) according to the manufacturer’s instructions. Cells were planted into 96-well plates at 5×103 cells/100 μL/well. After transfection, the optical density was measured at different points (0, 24, 48, and 72 hours) using a microplate reader (iMark; Bio-Rad Laboratories Inc., Hercules, CA, USA).
Migration and invasion assay
Cell migration and invasion abilities were assessed using the Transwell assay. For the invasion assay, the filters (8.0 μm pores, EMD Millipore) were additionally pre-coated with 30 μL 8× diluted matrix matrigel (BD Biosciences, San Jose, CA, USA) and dried at 37°C. Twenty four hours after transfection, cells were suspended in serum-free medium at a density of 1×105 per well and planted into the upper chamber. Complete medium (500 μL) with 10% fetal bovine serum was added to the lower chamber. After 48 hours of incubation at 37°C, the cells that adhered to the lower surface were fixed and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology). The cells were photographed and counted under a microscope.
Statistical analysis
Data are presented as the mean ± standard deviation (SD) from at least three independent experiments. Student’s t-test was used to compare the results. All statistical analyses were conducted using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). A P-value less than 0.05 was considered statistically significant.
Results
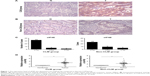
The upregulation frequency of eIF4E in the CLM group is higher than in the non-CLM group
A total of 80 pairs of tissue were included, and the cases were divided into the CLM group and non-CLM group (each group had 40 pairs of tissues). The levels of eIF4E in the paired tumor and nontumor tissues were detected by immunohistochemical staining and Western blot. The results of immunohistochemical staining are shown in Figure 1A and B. The eIF4E protein level was higher in tumor tissues than in matched nontumor tissues in both the CLM and non-CLM groups. Western blot indicated the frequency of eIF4E upregulation (ratio of tumor to nontumor tissue ≥2) was 82.5% (33/40) in the CLM group and 65.0% (26/40) in the non-CLM group (Figure 1C and D).
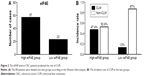
Upregulation of eIF4E in CRC patients is associated with liver metastasis
To clarify the prognostic value of eIF4E in CLM, we evaluated the level of eIF4E in tumor samples from 80 CRC patients. After surgical resection, the patients were divided into the high eIF4E expression group and low eIF4E expression group (ratio of tumor to nontumor tissue >2) according to Western blot analysis. Fifty-seven patients were included in the high eIF4E expression group, and 23 in the low eIF4E expression group (Figure 2A). After 2 years of follow-up, the results indicated the incidence rate of CLM in the high eIF4E expression group was 47.4% (27/57), while it was 13.0% in the low eIF4E expression group (3/23) (Figure 2B).
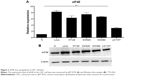
eIF4E is upregulated in CRC cell lines
To evaluate the expression level of eIF4E in CRC cells, RT-qPCR and Western blot analysis were performed. The five CRC cell lines (LS174T, HT-29, Lovo, SW480, and SW620) showed higher eIF4E expression levels than the normal tissue (three pooled normal tissues), and Lovo cells exhibited the highest eIF4E level (Figure 3A and B).
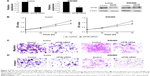
Downregulation of eIF4E inhibits proliferation, migration, and invasion of CRC cells
To explore the possible role of eIF4E in CRC progression, eIF4E-mediated effects on CRC cell proliferation, migration, and invasion were studied. eIF4E was knocked down using siRNA in Lovo and SW480 cells. Seventy two hours after transfection, RNA and protein were extracted, and qRT-PCR and Western blot were performed, respectively. The expression levels of eIF4E were reduced in both cell lines (Figure 4A). The CCK-8 assay indicated that knockdown of eIF4E inhibited cell growth compared with control, and the inhibition rate peaked at 72 hours (Figure 4B). The wound healing assay showed that downregulation of eIF4E suppressed the migratory activity of Lovo cells. Similar results were obtained in SW480 cells (Figure 4C). In the Transwell assay, cell invasion in two CRC cell lines was decreased following transfection with eIF4E siRNA, when compared to that in control cells (Figure 4D).
Knockdown of eIF4E suppresses the expression of cyclin D1, VEGF, MMP-2, and MMP-9 in CRC cells
To further explore the mechanism of eIF4E in regulating CRC metastasis, the protein levels of VEGF, MMP-2, and MMP-9 after eIF4E silencing were analyzed. siRNA-mediated knockdown of eIF4E decreased the cyclin D1, VEGF, MMP-2, and MMP-9 protein levels in Lovo (Figure 5A) and SW480 cell lines (Figure 5B).
Discussion
eIF4E has a critical role in facilitating mRNA translation. Accumulating evidence indicates that eIF4E is associated with cancer development and progression. Overexpression of eIF4E was observed in several cancer types such as breast cancer,16 gastric cancer,17 lung cancer,18 prostate cancer,19 and leukemia.20 High eIF4E expression level in breast cancer was associated with a greater rate of cancer recurrence and worse survival.16,21 Overexpression of eIF4E is associated with vascular invasion in gastric cancer,17 and it may serve as a prognostic and therapeutic factor for lung cancer, melanoma, and chronic lymphocytic leukemia (CLL) lymphocytes.20,22,23 In colorectal cancer, the eIF4E expression level increased steadily as the cancer progressed from benign dysplasia to adenocarcinoma.24 Therefore, eIF4E was selected for further research.
In the present study, the level of eIF4E in CRC tissues was evaluated, and the protein level of eIF4E was increased in the CLM and non-CLM tissues compared with matched nontumor tissues. This result is consistent with previous reports.24,25 The upregulation rate of eIF4E in the CLM group (82.5%) was much higher than that in the non-CLM group (65.0%), indicating that elevated eIF4E expression is associated with higher liver metastasis in CRC patients. We further confirmed the result by following 80 CRC patients after they underwent surgical resection. After 2 years, 27 patients in the high eIF4E expression group had liver metastasis, and only three patients in the low eIF4E expression group had liver metastasis; these results suggest that patients with high eIF4E expression level had a higher risk of liver metastasis. These data indicated eIF4E expression level is associated with liver metastasis in the CRC patients.
Further studies of eIF4E in CRC cells were performed. The eIF4E level was elevated in CRC cells compared to the normal tissue, and highly metastatic Lovo and SW620 cells have higher eIF4E levels than low metastatic LS174T, HT-29, and SW480 cells. We then investigated the effect of eIF4E on CRC metastasis by performing loss-of-function assays. Knockdown of eIF4E suppressed the proliferation, migration, and invasion of Lovo and SW480 cells. A major obstacle to CRC treatment is tumor metastasis, and the functional studies indicated that eIF4E is associated with tumor cell migration and invasion. As a result, we further explored the underlying mechanism.
Cyclin D1, VEGF, MMP-2, and MMP-9 are vital molecules involved in tumor metastasis.13 Cyclin D1 is required for cell growth and cell cycle control, and it is frequently overexpressed in human tumors.26 A previous study demonstrated that cyclin D1 was downregulated in eIF4E-silenced HeLa cells.27 The results of the present study showed that the expression of cyclin D1 was reduced after eIF4E knockdown in the two CRC cell lines. VEGF is a key mediator of tumor angiogenesis in the development of metastases.28 MMP-2 and MMP-9 are the MMPs that contribute to tumor invasion and metastasis by degrading collagen IV and other extracellular matrix proteins. The data indicated that with the downregulation of eIF4E, the expression levels of VEGF, MMP-2, and MMP-9 were reduced to a different extent. These results suggest that downregulation of eIF4E inhibits CRC cell metastasis partly through regulating the expression of cyclin D1, VEGF, MMP-2, and MMP-9.
Conclusion
In summary, the present study demonstrated that patients with high eIF4E expression levels had more frequent liver metastasis, suggesting the prognostic effect of eIF4E on CLM. Downregulation of eIF4E suppressed tumor metastasis, in part, through regulating the expression of cyclin D1, VEGF, MMP-2, and MMP-9.
Acknowledgments
The present study was funded by Science and Technology Development Project of Shandong Province (2014GSF118007) and Natural Science Foundation of Shandong Province (ZR2013HM061).
Disclosure
The author reports no conflicts of interest in this work.
References
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. | ||
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. | ||
Ueda M, Iguchi T, Nambara S, et al. Overexpression of Transcription Termination Factor 1 is Associated with a Poor Prognosis in Patients with Colorectal Cancer. Ann Surg Oncol. 2015;22(Suppl 3):1490–1498. | ||
Dahlmann M, Okhrimenko A, Marcinkowski P, et al. RAGE mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognostic biomarker for human colorectal cancer metastasis. Oncotarget. 2014;5(10):3220–3233. | ||
Jeong WJ, Cha PH, Choi KY. Strategies to overcome resistance to epidermal growth factor receptor monoclonal antibody therapy in metastatic colorectal cancer. World J Gastroenterol. 2014;20(29):9862–9871. | ||
Frankel TL, D’Angelica MI. Hepatic resection for colorectal metastases. J Surg Oncol. 2014;109(1):2–7. | ||
Lochan R, White SA, Manas DM. Liver resection for colorectal liver metastasis. Surg Oncol. 2007;16(1):33–45. | ||
Truitt ML, Conn CS, Shi Z, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162(1):59–71. | ||
Zhang B, Dong S, Zhu R, et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget. 2015;6(26):22799–22811. | ||
Carroll M, Borden KL. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res. 2013;33(5):227–238. | ||
Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. | ||
Nagaraju GP, Bramhachari PV, Raghu G, El-Rayes BF. Hypoxia inducible factor-1alpha: Its role in colorectal carcinogenesis and metastasis. Cancer Lett. 2015;366(1):11–18. | ||
Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16(20):4914–4920. | ||
Xu T, Zhou M, Peng L, et al. Upregulation of CD147 promotes cell invasion, epithelial-to-mesenchymal transition and activates MAPK/ERK signaling pathway in colorectal cancer. Int J Clin Exp Pathol. 2014;7(11):7432–7441. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. | ||
Heikkinen T, Korpela T, Fagerholm R, et al. Eukaryotic translation initiation factor 4E (eIF4E) expression is associated with breast cancer tumor phenotype and predicts survival after anthracycline chemotherapy treatment. Breast Cancer Res Treat. 2013;141(1):79–88. | ||
Liang S, Guo R, Zhang Z, et al. Upregulation of the eIF4E signaling pathway contributes to the progression of gastric cancer, and targeting eIF4E by perifosine inhibits cell growth. Oncol Rep. 2013;29(6):2422–2430. | ||
Li Y, Fan S, Koo J, et al. Elevated expression of eukaryotic translation initiation factor 4E is associated with proliferation, invasion and acquired resistance to erlotinib in lung cancer. Cancer Biol Ther. 2012;13(5):272–280. | ||
Graff JR, Konicek BW, Lynch RL, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69(9):3866–3873. | ||
Martinez-Marignac V, Shawi M, Pinedo-Carpio E, et al. Pharmacological targeting of eIF4E in primary CLL lymphocytes. Blood Cancer J. 2013;3:e146. | ||
Flowers A, Chu QD, Panu L, et al. Eukaryotic initiation factor 4E overexpression in triple-negative breast cancer predicts a worse outcome. Surgery. 2009;146(2):220–226. | ||
Wang R, Geng J, Wang JH, Chu XY, Geng HC, Chen LB. Overexpression of eukaryotic initiation factor 4E (eIF4E) and its clinical significance in lung adenocarcinoma. Lung Cancer. 2009;66(2):237–244. | ||
Khosravi S, Tam KJ, Ardekani GS, Martinka M, McElwee KJ, Ong CJ. eIF4E is an adverse prognostic marker of melanoma patient survival by increasing melanoma cell invasion. J Invest Dermatol. 2015;135(5):1358–1367. | ||
Diab-Assaf M, Abou-Khouzam R, Saadallah-Zeidan N, et al. Expression of eukaryotic initiation factor 4E and 4E binding protein 1 in colorectal carcinogenesis. Int J Clin Exp Pathol. 2015;8(1):404–413. | ||
Rosenwald IB, Chen JJ, Wang S, Savas L, London IM, Pullman J. Upregulation of protein synthesis initiation factor eIF-4E is an early event during colon carcinogenesis. Oncogene. 1999;18(15):2507–2517. | ||
Ichikawa M, Sowa Y, Iizumi Y, Aono Y, Sakai T. Resibufogenin induces G1-phase arrest through the proteasomal degradation of cyclin D1 in human malignant tumor cells. PloS One. 2015;10(6):e0129851. | ||
Othumpangat S, Kashon M, Joseph P. Sodium arsenite-induced inhibition of eukaryotic translation initiation factor 4E (eIF4E) results in cytotoxicity and cell death. Mol Cell Biochem. 2005;279(1–2):123–131. | ||
Chen F, Wang M, Bai J, et al. Role of RUNX3 in suppressing metastasis and angiogenesis of human prostate cancer. PloS One. 2014;9(1):e86917. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.