Back to Journals » OncoTargets and Therapy » Volume 10
Overexpression of ecotropic viral integration site-1 is a prognostic factor of lung squamous cell cancer
Received 14 January 2017
Accepted for publication 17 March 2017
Published 26 May 2017 Volume 2017:10 Pages 2739—2744
DOI https://doi.org/10.2147/OTT.S132410
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Xueliang Xu,1 Shengchen Liu,2 Xia Ji1
1Department of Respiratory Medicine, 2Department of Emergency, Linyi People’s Hospital, Linyi, China
Aim: To explore the expression and clinical significance of ecotropic viral integration site-1 (EVI1) of lung squamous cell cancer (SCC).
Methods: The expression of EVI1 in SCC was detected by immunohistochemistry and the validation cohort was divided into EVI1 high-expression group and low-expression group according to the cutoff of immunohistochemical score. The correlations between EVI1 expression and the clinicopathological factors were analyzed by χ2 test. The relation between EVI1 expression and overall survival rate was evaluated by univariate analysis with Kaplan–Meier method. The independent prognostic factor was identified by multivariate analysis with Cox regression model.
Results: In this study, the EVI1 high-expression percentage was 32.32% (53/164). EVI1 high expression was significantly associated with a poorer overall 5-year survival rate of SCC (P=0.021). Moreover, EVI1 high expression was identified as an independent prognostic factor of SCC, predicting the unfavorable prognosis (P=0.013).
Conclusion: High expression of EVI1 was significantly associated with a poorer prognosis and it was identified as an independent prognostic factor of SCC.
Keywords: ecotropic viral integration site-1, lung squamous cell cancer, biomarker, prognosis, immunohistochemistry, overall survival rate
Introduction
Lung cancer is becoming a more and more critical public health problem and medical insurance burden, resulting in the largest number of cancer-related deaths worldwide.1 Approximately 1,590,000 patients die of lung cancer in the world every year.2 Due to severe air pollution, the mortality and morbidity of lung cancer are much more critical in developing countries like China and India. Pathologically, non-small cell lung cancer (NSCLC) takes up ~80%–85% in lung cancer, including two main subtypes, the adenocarcinoma (AC) and squamous cell cancer (SCC). The former type occupies ~40% and the latter type accounts for 25%–30% in all kinds of lung cancers.3 Although surgical techniques and adjuvant therapies have improved remarkably, the 5-year overall survival rate of lung cancer is still very unsatisfactory, remaining 10%–15%.1 There is an urgent need for effective biomarkers of lung cancer, because of the ability to help identify high-risk patients, facilitate the application of optimal treatment, and improve the overall survival rate.
Ecotropic viral integration site-1 (EVI1) is identified as an oncoprotein mostly in hematological malignancies such as acute myelogenous leukemia (AML) and myelodysplastic syndrome (MDS). Frequent activation of EVI gene could be observed for intrachromosomal or interchromosomal rearrangements in patients with AML or MDS. EVI1 was also demonstrated to regulate lots of cellular processes in tumorigenesis and progression, including cell proliferation, transformation, differentiation, and survival, especially in leukemic cells.4–8 In solid tumors, such as glioblastoma and prostate cancer, EVI1 was also reported to be involved in progression or prognosis.7,9 In SCC, the amplification of chromosome 3q26.2–q29 was frequently observed.10 This region (3q26.2–q29) included the location of EVI1 gene, indicating the possible oncogenic role of EVI1 in lung cancer, especially in SCC. But that study focused on the gene level and could not provide the direct evidence of EVI1 protein as an oncoprotein in SCC.10 In conclusion, the expression, intracellular location, and function of EVI1 have not been explored in SCC.
Although SCC and AC are included in NSCLC with a similar treatment strategy and outcome, their biological features are different.11 Based on the previous indication that the region containing EVI1 gene was amplified in SCC,10 it was suspected that EVI1 might be involved in SCC progression or prognosis. Therefore, the expression of EVI1 was detected in 164 cases of SCC with immunohistochemistry (IHC) and the clinical and prognostic significance of EVI1 in SCC was evaluated further.
Patients and methods
Patients and follow-ups
This study was reviewed and approved by the Ethics Committee of Linyi People’s Hospital. The primary cohort consisted of 461 patients diagnosed with SCC in the medical center of Linyi People’s Hospital, and the validation cohort was selected from the primary cohort based on the following criteria: 1) available paraffin-embedded tissues; 2) no adjuvant chemotherapy or radiation therapy before or after surgery; and 3) radical section of the tumor. According to the above criteria, a total of 164 SCC patients were selected for the validation cohort. All the samples were obtained from the pathology department with prior written informed consent of patients’ family for this study. The tissue samples were sliced and stained with hematoxylin-eosin for double confirmation of diagnosis and selection of IHC area. The pathological tumor-node-metastasis (pTNM) stage of SCC was identified by the seventh American Joint Committee on Cancer/Union for International Cancer Control staging system (2009).
IHC
The protocol of IHC staining was performed according to previous studies.12,13 Briefly, the paraffin-embedded SCC samples were deparaffinized with xylene and graded ethanol. Then the slides were incubated in citrate buffer for 30 minutes for antigen retrieval. Blockage of endogenous peroxidase enzyme was achieved by incubation in 3% hydrogen peroxide in methanol for 20 minutes. Primary anti-EVI1 antibody (Novus Biologicals, Littleton, CO, USA) with dilution at a ratio of 1:100 was used to incubate the tissues at 4°C overnight, followed by a wash of phosphate-buffered saline. The corresponding biotinylated secondary antibody and streptavidin-peroxidase complex were used to incubate at 37°C for 30 minutes. Finally, the slides were visualized by incubation with 3,3′-diaminobenzidine solution.
Evaluation of IHC results
According to previous studies, the IHC results were evaluated by the IHC score, which was defined as the product of score of positive cell percentage and the score of staining intensity.14,15 The score of positive cell percentage was defined as follows: 0 for 10% positive cells; 1 for 10%–30% positive cells; 2 for 30%–50% positive cells; and 3 for ≥50% positive cells. The score of staining intensity was described as follows: 0 for negative staining; 1 for weak staining; 2 for moderate staining; and 3 for strong staining. The total score of IHC was defined as the product of score for staining intensity and the score for positive cells. The total IHC score ranged from 0 to 9, and the cutoff was selected with a receiver operating characteristic curve with the highest sensitivity plus specificity.16 The cohort was divided into EVI1 high-expression group and EVI1 low-expression group, according to the cutoff of EVI1 IHC score.
Statistical analysis
SPSS 17.0 software (IBM Corp., Armonk, NY, USA) was used to analyze all the data. The χ2 test was used to analyze the correlation between EVI1 expression and the clinicopathological factors. The Kaplan–Meier method was performed for the survival curve and the log-rank test was used to compare the statistical difference between the subtypes. The Cox regression hazard model was used for multivariate analysis. A P-value of <0.05 was considered to be statistically significant.
Results
Information of validation cohort
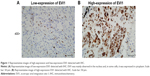
A total of 164 patients diagnosed with SCC were enrolled into our cohort, including 106 male and 58 female patients (Table 1). The average age of patients was 64.5 years. The median postoperational survival time was 22 months and the average survival time was 34 months. As previously described in the “Patients and methods” section, the cohort was divided into EVI1 high-expression group and EVI1 low-expression group according to the cutoff of EVI1 IHC score. In this study, the percentage of EVI1 high-expression group was 32.32% (53/164). And EVI1 was mainly observed in the nucleus of SCC cells, but in some cases it was also expressed in cytoplasm. Representative images of low-expression and high-expression EVI1 are displayed in Figure 1.
  | Table 1 Characteristics of patients with SCC |
Correlation between the clinicopathological factors and EVI1 expression
After the cohort was divided into EVI1 high-expression group and EVI1 low-expression group, the correlation between EVI1 expression and other clinicopathological factors was analyzed by χ2 test. The clinicopathological factors included the age, gender, lymphatic status, tumor size, smoking status, tumor differentiation, and pTNM stage of patients (Table 2). Considering that the patients of stage IV already have a distant metastasis and the radical surgery cannot be achieved as usual, the pTNM stage only included stages I–III in this cohort. The analyses of correlation between the clinicopathological factors and EVI1 expression could help find the potential association of EVI1 with the progression of SCC. However, there was no significant clinicopathological factor identified to be significantly associated with EVI1 expression in this study.
  | Table 2 Correlation between EVI1 expression and the clinicopathological parameters |
Prognostic value of EVI1 in SCC
In previous study,10 the expression of EVI1 was demonstrated to be much higher in SCC compared with AC. It was suspected that EVI1 expression might affect the prognosis of SCC and therefore its prognostic significance was evaluated with univariate and multivariate analyses (Table 3). The univariate analysis with Kaplan–Meier method was performed first to analyze the relation between EVI1 expression and the overall 5-year survival rate. In the univariate analysis, positive lymphatic invasion (P<0.001, 5-year survival rate: 13.0 vs 38.4 months) and advanced TNM stage (P<0.001, 5-year survival rate: 0 vs 22.5 vs 32.4 months) were proved to be significantly associated with the poorer prognosis of SCC (Figure 2A and B). Notably, the high expression of EVI1 could also predict the unfavorable prognosis of SCC (P=0.021, 18.5 vs 26.6 months) (Figure 2C). No other clinicopathological factor was defined as the prognostic factor in this study.
The multivariate analysis with the Cox regression model was further performed to detect the independent prognostic factors. All the clinicopathological factors were enrolled into the Cox regression model except TNM stage because of its obvious interaction with lymphatic status. In the multivariate analysis, the high expression of EVI1 was identified as an independent prognostic factor of SCC (hazard ratio [HR] =1.66, 95% confidence interval [CI] =1.11–2.47, P=0.013), indicating that the high expression of EVI1 alone was enough to predict the poorer prognosis of SCC. Besides EVI1, lymphatic status was also defined as an independent prognostic factor in this study (HR =2.45, 95% CI =1.60–3.76, P<0.001).
Discussion
SCC and AC are always classified into one type-NSCLC because of their similar treatment strategy. However, they are different histological types and have significantly different phenotypic properties. In previous studies, the most valuable study inspiring us to carry out the experiments of this study was conducted by Kang et al.10 They performed high-resolution array-comparative genomic hybridization and identified the differences in the patterns of genomic imbalances between SCC and AC. As a result, they demonstrated that SCC instead of AC has an amplified region of gene in 3q26.2–q29, where the EVI1 gene is located.10 Many previous studies have proved the oncogenic role of EVI1 in tumors, especially tumors of hematopoietic system.17 More and more emerging evidence has indicated the oncogenic role of EVI1 in solid tumors, including prostate cancer, ovarian cancer, glioblastoma, hepatocellular carcinoma, and so on.7,9,18 The underlying molecular mechanism of how EVI1 promotes cancer progression and affects prognosis has not been well elucidated. Yasui et al proved that EVI1 could antagonize transforming growth factor-β-mediated growth inhibition in hepatocellular carcinoma, which promotes the proliferation of hepatocellular carcinoma cells.18 However, some sporadic studies pointed out the dual role of EVI1 in cancer progression. For example, Nanjundan et al19 pointed out that amplification of EVI1 is associated with a favorable prognosis in ovarian cancer. Although that study was based on the detection of DNA copy number, it still reflects the controversial role of EVI1 in some special cancer types.19 In conclusion, many studies20,21 identified EVI1 as a protein promoting cancer progression, but consensus has not been reached. More experiments need to be performed for validation if new cancer types are involved, such as SCC.
Nowadays, the trend of the precise treatment is becoming more and more prevalent. The theoretical basis of the precise treatment is the individual heterogeneity and diversity of patients. Different cancers may exhibit different varieties and abundance of oncogenes or oncoproteins. In postoperational patients with cancer, detection of these oncogenes or oncoproteins is the first step to direct chemotherapy or target therapy. A large number of predicting or prognostic biomarkers have been explored in lung cancer, but only a few have been demonstrated to be clinically relevant. The unionization of medical centers is necessary to enlarge the sample size and reduce diversity, which would help in getting more solid results. Moreover, the detection of candidate proteins in situ may be more valuable for prediction. Detecting the expression of proteins in tumor samples directly could reduce the confounding factors from gene modification to messenger RNA transcription. In tumor treatment therapy, the finding of an effective biomarker could be revolutionary, leading to a targeted drug, or even a potential therapy, just like the exploration of human epidermal growth factor receptor 2 (HER2) function in breast cancer resulted in the development of trastuzumab and the target therapy to patients with breast cancer. The finding of this study that the high expression of EVI1 is an independent prognostic factor could help more specifically stratify SCC high-risk and low-risk patients and help them get different treating therapies. Moreover, along with the elucidation of molecular mechanisms of how EVI1 leads to the poorer prognosis of SCC, EVI1 may become a potent drug target, like the revolution triggered by HER2 in breast cancer treatment. The finding of clinical significance of EVI1 in SCC could inspire more interest on EVI1 and encourage more experiments to explore the underlying mechanisms of EVI1 downstream pathways.
In conclusion, the expression of EVI1 was detected for the first time in 164 cases of SCC samples with IHC, and the cohort was divided into EVI1 high-expression and low-expression groups according to the score. Furthermore, it was demonstrated that the high expression of EVI1 was significantly associated with the unfavorable prognosis of SCC and that EVI1 was an independent prognostic factor of SCC. The findings could trigger interest in the study on the molecular mechanism of EVI1 as an oncoprotein in SCC and help identify the substrate regulated by EVI1. The results could help more specifically stratify SCC high-risk and low-risk patients and help find a potential drug target in SCC treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. | ||
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 6. 2015. J Natl Compr Canc Netw. 2015;13(5):515–524. | ||
Nucifora G, Laricchia-Robbio L, Senyuk V. EVI1 and hematopoietic disorders: history and perspectives. Gene. 2006;368:1–11. | ||
Heller G, Rommer A, Steinleitner K, et al. EVI1 promotes tumor growth via transcriptional repression of MS4A3. J Hematol Oncol. 2015;8:28. | ||
Wang H, Schaefer T, Konantz M, et al. Prominent oncogenic roles of EVI1 in breast carcinoma. Cancer Res. 2017;77:2148–2160. | ||
Queisser A, Hagedorn S, Wang H, et al. Ecotropic viral integration site 1, a novel oncogene in prostate cancer. Oncogene. 2017;36:1573–1584. | ||
Sato T, Goyama S, Kataoka K, et al. Evi1 defines leukemia-initiating capacity and tyrosine kinase inhibitor resistance in chronic myeloid leukemia. Oncogene. 2014;33(42):5028–5038. | ||
Hou A, Zhao L, Zhao F, et al. Expression of MECOM is associated with unfavorable prognosis in glioblastoma multiforme. Onco Targets Ther. 2016;9:315–320. | ||
Kang JU, Koo SH, Kwon KC, Park JW, Kim JM. Identification of novel candidate target genes, including EPHB3, MASP1 and SST at 3q26.2-q29 in squamous cell carcinoma of the lung. BMC Cancer. 2009;9:237. | ||
Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388(10048):1012–1024. | ||
Xu YF, Yang XQ, Lu XF, et al. Fibroblast growth factor receptor 4 promotes progression and correlates to poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;446(1):54–60. | ||
Xu Y, Yang X, Li Z, et al. Sprouty2 correlates with favorable prognosis of gastric adenocarcinoma via suppressing FGFR2-induced ERK phosphorylation and cancer progression. Oncotarget. 2017;8:4888–4900. | ||
Xu YF, Ge FJ, Han B, et al. High-mobility group box 1 expression and lymph node metastasis in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2015;21(11):3256–3265. | ||
Liu H, Zhang Q, Li K, et al. Prognostic significance of USP33 in advanced colorectal cancer patients: new insights into β-arrestin-dependent ERK signaling. Oncotarget. 2016;7:81223–81240. | ||
Liu H, Xu Y, Zhang Q, et al. Correlations between TBL1XR1 and recurrence of colorectal cancer. Sci Rep. 2017;7:44275. | ||
Fenouille N, Bassil CF, Ben-Sahra I, et al. The creatine kinase pathway is a metabolic vulnerability in EVI1-positive acute myeloid leukemia. Nat med. Epub 2017 Feb 13. | ||
Yasui K, Konishi C, Gen Y, et al. EVI1, a target gene for amplification at 3q26, antagonizes transforming growth factor-β-mediated growth inhibition in hepatocellular carcinoma. Cancer Sci. 2015;106(7):929–937. | ||
Nanjundan M, Nakayama Y, Cheng KW, et al. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007;67(7):3074–3084. | ||
Hinai AA, Valk PJ. Review: Aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol. 2016;172(6):870–878. | ||
Wieser R. The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene. 2007;396(2):346–357. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.