Back to Journals » Cancer Management and Research » Volume 12
Overexpression of Cancer Upregulated Gene 2 (CUG2) Decreases Spry2 Through c-Cbl, Leading to Activation of EGFR and β-Catenin Signaling
Authors Yawut N , Kaewpiboon C, Budluang P, Cho IR , Kaowinn S , Koh SS, Chung YH
Received 10 July 2020
Accepted for publication 9 September 2020
Published 15 October 2020 Volume 2020:12 Pages 10243—10250
DOI https://doi.org/10.2147/CMAR.S271109
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
Natpaphan Yawut,1,* Chutima Kaewpiboon,2,* Phatcharaporn Budluang,1 Il-Rae Cho,1 Sirichat Kaowinn,3 Sang Seok Koh,4 Young-Hwa Chung1
1BK21 Plus, Department of Cogno-Mechatronics Engineering, Pusan National University, Busan 46241, Republic of Korea; 2Department of Biology, Faculty of Science, Thaksin University, Pattalung 93210, Thailand; 3Department of General Science and Liberal Arts, King Mongkut’s Institute of Technology, Ladkrabang Prince of Chumphon Campus, Chumphon 86160, Thailand; 4Department of Biosciences, Dong-A University, Busan 49315, Republic of Korea
*These authors contributed equally to this work
Correspondence: Young-Hwa Chung
Department of Cogno-Mechatronics Engineering, Pusan National University, Geumjeong Gu, Busan Daehak Ro 63-2, Busan 46241, Republic of Korea
Email [email protected]
Purpose: The mechanism by which cancer upregulated gene 2 (CUG2) overexpression induces cancer stem cell-like phenotypes is not fully understood. Because the increased activity and expression of epidermal growth factor receptor (EGFR) kinase have been reported in A549 cancer cells overexpressing CUG2 (A549-CUG2) compared with control cells (A549-Vec), the Sprouty2 (Spry2) protein has gained attention as the downstream molecule of EGFR signaling. Therefore, we aim to identify the role of Spry2 in CUG2-overexpressing lung cancer cells.
Materials and Methods: Spry2 expression levels were examined in A549-CUG2 and A549-Vec cells by Western blotting and qRT-PCR. Cell migration, invasion, and sphere formation were examined after Spry2 suppression and overexpression. EGFR-Stat1 and Akt-ERK protein phosphorylation levels were detected via immunoblotting. NEK2 kinase and β-catenin reporter assay were performed for downstream of Spry2 signaling.
Results: Although A549-CUG2 cells showed lower levels of the Spry2 protein than A549-Vec cells, no difference in levels of Spry2 transcript was observed between both cells via qRT-PCR. Furthermore, MG132 treatment enhanced the protein levels and ubiquitination of Spry2, suggesting that Spry2 protein expression can be regulated via the ubiquitin-proteasome pathway. The enforced expression of c-Cbl, known as the binding partner of Spry2, decreased the Spry2 protein levels, whereas its knockdown oppositely increased them. Epithelial–mesenchymal transition (EMT) and sphere formation were increased in A549-Vec cells during Spry2 siRNA treatment, confirming the role of Spry2 in CUG2-induced oncogenesis. Furthermore, EMT and sphere formation were determined by the Spry2 protein levels through the regulation of EGFR-Stat1 and β-catenin-NEK2-Yap1 signaling pathways.
Conclusion: CUG2 reduces Spry2 protein levels, the negative signaling molecule of cell proliferation, via c-Cbl, possibly activating the EGFR and β-catenin signaling pathways and, in turn, contributing to the induction of cancer stem cell-like phenotypes.
Keywords: CUG2, cancer stem cell-like phenotypes, Spry2, c-Cbl
Introduction
Cancer upregulated gene 2 (CUG2) is suggested to have oncogenic activity as Affymetrix microarray has shown that it is upregulated in tumors of several tissues such as lungs, ovaries, and colon, resulting in tumor formation in nude mice due to the overexpression of the protein.1 Recent studies have reported that CUG2 overexpression induces cancer stem cell-like phenotypes including the increase in cell migration, invasion, sphere formation, and resistance to anticancer drugs via the upregulation of TGF-β signaling.2–4 Furthermore, our studies have revealed that EGFR-Stat1-HDAC4 signaling axis and β-catenin-NEK2-Yap1 signaling axis are also involved in CUG2-induced cancer stem cell-like phenotypes.3,5
After Sprouty proteins were first reported to be involved in the inhibition of FGF- or EGF-induced signaling during Drosophila development,6,7 detailed action mechanisms of the protein have been revealed. For example, Sprouty 2 (Spry2) hinders Grb2-SOS interaction and Raf activation.8,9 However, some studies have shown that Spry2 sustains EGFR signaling by interacting with c-Cbl, an E3 ubiquitin ligase, resulting in the inhibition of EGFR degradation.10,11 Clinical evidence suggests that decreased levels of Spry2 are detected in lung,12 breast,13 and prostate cancer.14 On the other hand, elevated levels of Spry2 are found in KRasG12V-expressing transgenic mice,15 where it participates in a negative feedback mechanism; leading to the suppression of lung tumorigenesis.
The product of the mammalian c-Cbl gene is widely expressed in the cytoplasm of the cell and is involved in the negative regulation of receptor tyrosine kinase (RTK) signaling via RTK ubiquitination, subsequently leading to the degradation of the RTK.16,17 Supporting this function, the domain structure of c-Cbl contains a tyrosine- kinase-binding domain, a RING finger domain, and a C-terminal ubiquitin-associated domain.18 The downregulation of Spry2 mediated by c-Cbl E3 ligase has been reported during EGF or FGF signaling.19
This study was initiated to investigate more detailed mechanisms of CUG2-induced oncogenesis. We herein report that CUG2 represses Spry2 via c-Cbl E3 ligase, which upregulates EGFR and β-catenin signaling. Eventually, the activation of these signaling pathways contributes to the induction of EMT and sphere formation, which are cancer stem cell-like phenotypes.
Materials and Methods
Cell Culture
Human lung cancer A549 cells were obtained from American Type Culture Collection (Manassas, VA, USA). A549 cancer cells stably expressing CUG2 (A549-CUG2) and an empty vector (A549-Vec) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS, penicillin, and streptomycin under G410 (500 µg/mL).
Antibodies and Reagents
Antibodies against β-catenin, Akt, ERK, and Stat1, and its phosphorylated proteins were purchased from Cell Signaling Biotechnology (Danvers, MA, USA). Antibodies against E-cadherin, -N-cadherin, -vimentin, -NEK2, and -Yap1 were obtained from Abcam (Cambridge, MA, USA). Anti-Spry2 and -c-Cbl antibodies were acquired from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell Transfection
Cells were transfected with Spry2 siRNA, c-Cbl siRNA, pcDNA3-Cbl, or pHM6-Spry2 vector (provided from Prof. Tarun B. Patel, Albany College of Pharmacy and Health Sciences, NY, USA) with Lipofectamine 2000 at 80% confluence and harvested for immunoblotting at 40 h post-transfection.
Wound Healing Assay
An artificial wound was generated on a confluent cell monolayer using a 200 µl of pipette tip. The cells were then incubated for 24 h to measure the closure of the scratch.
Transwell Invasion Assay
Cells (1 x 104 cells/well) were seeded in the upper well containing DMEM without the serum and further cultured for 24 h to allow the cells to migrate into the lower well containing the serum through a membrane coated with Matrigel (BD Bioscience, San Jose, CA, USA). The migrated cells were counted after fixation with 4% paraformaldehyde and staining with eosin for 30 min.
Sphere Forming Assay
Cells were incubated in 24-well ultralow attachment plates containing serum-free medium supplemented with insulin (5 µg/mL), BSA (0.4%), basic FGF (10 ng/mL), and EGF (20 ng/mL) for 6 days. The size and number of spheroids were analyzed under a light microscope (CKX31-11 PHP, Olympus, Japan).
Immunoblotting and Immunoprecipitation
Total proteins were extracted from cells lysed with RIPA buffer containing a protease inhibitor cocktail and were then separated using 10% SDS-PAGE. After the transfer of the gel to a PVDF membrane, the membrane was incubated with primary antibodies (1:500 dilution) and subsequently with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:1,000 dilution). The blot was developed by the addition of ECL solution, and then imaging was acquired using ImageQuant LAS 4000 Mini (GE-Healthcare, Tokyo, Japan). For immunoprecipitation, cells were harvested after 48 h of transfection, and the cell debris was removed by centrifugation at 10,000 g for 10 min at 4°C. After preclearance with 25 µL of protein A/G agarose, cell lysates were incubated with an appropriate primary antibody and protein A/G agarose for 1 h at 4°C. The precipitates were resolved on SDS-PAGE gels and analyzed by immunoblotting with appropriate antibodies.
Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from cells, and the cDNAs were synthesized using a QuantiTect Probe reverse transcriptase-PCR (RT-PCR) Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instructions. Real-time RT-PCR was conducted on a SYBR Premix Exicycler 96 Real-Time Quantitative Thermal Block (Bioneer. Daejeon, Korea) with the following parameters: 40 cycles for 95°C for 5 sec, 58°C for 25 sec and 72°C for 30 sec. The reaction mixture for the real-time PCR was assembled with 25µl of AccuPower 2x GreenStar Master Mix (Bioneer), 19 µl of deionized sterile water, 5 µl of sense and antisense primers, and 1 µl of cDNA in 50 µl of total volume. The real-time RT-PCR data were obtained in the form of threshold cycle (Ct) values, and target gene expression was normalized to a GADPH expression. Relative expression levels of the target gene (Spry2) were calculated by the comparative Ct (2−ΔΔCτ) method as previously described.20,21
Immunofluorescence Microscopy
For performing immunofluorescence microscopy, cells grown on coverslips were fixed with 4% paraformaldehyde for 15 min, permeabilized with cold acetone for 15 min, blocked with 10% goat serum for 30 min, and treated with anti-Spry2 antibodies for 30 min at room temperature. Then, the cells were washed and incubated with Alexa Fluor 488-conjugated goat anti-rabbit antibody (A11008) in PBS for 30 min and washed three times with PBS. For performing nuclear staining, the cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min in the dark and were washed three times using PBS. Subsequently, coverslips with stained cells were mounted on slides using PBS containing 10% glycerol and were imaged using a fluorescence microscope (Zeiss Axio Observer D1, Oberkochen, Germany).
Luciferase Reporter Assay
The A549-CUG2 cells were transfected with Top-Flash, or Fop-Flash vectors using Lipofectamine 2000. To normalize transfection efficiency, a pGK-βgal vector expressing β-galactosidase under the control of a phosphoglucokinase promoter was included in the transfection mixture. At 48 h after transfection, the cells were washed with cold PBS and lysed in a lysis solution (25 mM Tris [pH 7.8], 2 mM EDTA, 2 mM DTT, 10% glycerol, and 1% TritonX-100), and luciferase activity was measured using a luminometer and a luciferase kit (Promega, Madison, WI, USA).
Statistical Analysis
Data were presented as means ± standard deviation (SD). To compare the difference between two groups, an unpaired t-test was used with the GraphPad Prism software. P-value of <0.05 was considered statistically significant.
Results
CUG2 Overexpression Decreases Spry2 Expression via c-Cbl E3 Ligase
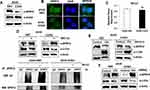
As our previous studies revealed elevated EGFR expression and activation of downstream signaling pathways such as Akt-ERK pathway in A549-CUG2 cells,22,23 we paid attention to the status of Spry2, a regulatory molecule involved in EGFR signaling. We then found that Spry2 protein levels were lower in A549-CUG2 cells than in A549-Vec cells as a control (Figure 1A). We confirmed that Spry2 was much less stained with immunofluorescence in A549-CUG2 cells compared with A549-Vec cells (Figure 1B). To examine whether the lower levels of the Spry2 protein in A549-CUG2 cells were attributed to low Spry2 transcript levels, we performed qRT-PCR in both A549-CUG2 and A549-Vec cells after total RNA isolation using an RNA miniprep kit. As shown in Figure 1C, Spry2 transcript levels in A549-CUG2 cells were similar to that in A-549-Vec cells. Furthermore, because it has been reported that Spry2 protein levels are also regulated at the post-transcriptional level such as the ubiquitin-proteasome pathway, treatment with MG132, the inhibitor of proteasome, enhanced protein levels of Spry2 in both the cells. We also observed enhanced ubiquitination of Spry2 under MG132 treatment (Figure 1D). As other studies have reported that Spry2 interacts with and is the target of c-Cbl E3 ligase,24,25 we introduced c-Cbl expression vector or c-Cbl siRNA into both cells. As shown in Figure 1E and F, the enforced expression of c-Cbl decreased Spry2 protein levels, whereas its knockdown enhanced protein levels. These results suggest that CUG2 overexpression induces the degradation of Spry2 protein via c-Cbl E3 ligase, the downstream molecule of EGFR signaling.
Spry2 Protein Levels Determine the Capacity of EMT and Sphere Formation
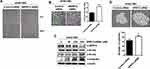
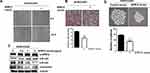
To explore whether Spry2 actually affects CUG2-induced cancer stem cell-like phenotypes, we reduced the Spry2 protein levels using Spry2 siRNA in A549-Vec cells. Thereafter, we examined the capability of EMT and sphere formation in A549-Vec cells. We then found that Spry2 siRNA treatment enhances cell migration and invasion in A549-Vec cells (Figure 2A and B). These results were further supported by detecting the decreased level of E-cadherin, and the increased level of N-cadherin and vimentin after Spry2 siRNA treatment (Figure 2C), depending on the Spry2 siRNA concentration. Spry2 suppression increased sphere size and number (Figure 2D). On the other hand, we attempted the enforced expression of Spry2 by introducing Spry2-expressing vector into A549-CUG2 cells. We found that the enforced expression of Spry2 inhibited CUG2-induced cell migration and invasion (Figure 3A and B), which are opposite to the results of Spry2 siRNA treatment. The enforced expression of Spry2 increased E-cadherin expression, and decreased N-cadherin and vimentin protein levels (Figure 3C), which can explain the inhibition of EMT in A549-CUG2 cells. The transient introduction of the Spry2-expressing vector into A549-CUG2 cells significantly diminished sphere size and number (Figure 3D). Taken together, we suggest that Spry2 protein levels can determine the capacity of EMT and sphere formation in A549 lung cancer cells.
The Levels of Spry2 Determine the Activation of EGFR-Stat1–HDAC4 and β-Catenin-NEK2-Yap1 Signaling Pathways
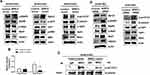
We observed that the enforced expression of Spry2 inhibited EMT and sphere formation (Figure 3A-D); however, the mechanism by which Spry2 affects these cancer stem cell-like phenotypes was not yet studied. Based on our previous research, we believe that the EGFR-Stat1-HDAC4 and β-catenin-NEK2-Yap1 signaling pathways are involved in cancer stem cell-like phenotypes;3,5 however, whether Spry2 hinders these pathways has not yet been investigated. To test our hypothesis, we examined the levels of these signaling proteins under Spry2 overexpression. As shown in Figure 4A, the enforced expression of Spry2 reduced phospho-EGFR, and phosphorylation levels of its downstream molecules such as Stat1, Akt, and ERK also decreased. HDAC4 protein levels were also reduced after the introduction of the Spry2 expression vector into A549-CUG2 cells (Figure 4A). In addition, when we examined the β-catenin–NEK2-Yap1 signaling pathway under the enforced Spry2 condition, we found that there was decrease in phospho-β-catenin (Se34/37), NEK2, and phospho-Yap1 (Y357) levels (Figure 4A). Moreover, we observed diminished β-catenin transcriptional activity and NEK2 kinase activity (Figure 4B and C). Inversely, Spry2 knockdown increased the phosphorylation levels of EGFR, Stat1, β-catenin (Se34/37), and Yap1 (Y357) (Figure 4D). HDAC4 expression was also increased during Spry2 siRNA treatment (Figure 4D). In addition, Spry2 suppression increased EGFR, Stat1, β-catenin, and NEK2 protein levels (Figure 4D). Interestingly, we found that Spry2 siRNA treatment decreased Yap1 protein levels (Figure 4D). These results suggest that Spry2 levels determine the activation of EGFR-Stat1-HDAC4 and β-catenin–NEK2-Yap1 signaling pathways.
Discussion
The mechanism by which CUG2 overexpression maintains the activation of EGFR- and β-catenin-mediated signaling remains unanswered. Therefore, we paid attention to regulatory molecules of EGFR downstream signaling, and attempted to examine the status of Spry2 protein, the downstream regulatory molecule of EGFR signaling.8,26 We found low Spry2 protein levels in A549-CUG2 cells due to E3 ligase c-Cbl-mediated ubiquitination, indicating the post-transcriptional regulation of Spry2 under CUG2 overexpression. Interestingly, the enforced expression of Spry2 hindered cell migration, invasion, and sphere formation, whereas its knockdown resulted in the acceleration of cancer stem cell-like phenotypes, suggesting the role of Spry2 as a tumor suppressor. Similarly, other studies showing that Spry2 protein levels determine cell migration and invasion in pancreatic and gastric cancer cells27,28 also supported our results. Considering the known regulatory role of Spry2 in RTK signaling,6,7 Spry2 overexpression reduces EGFR-Stat1-HDAC4 and Akt-ERK signaling, whereas its knockdown shows opposite results.
As another target signaling of Spry2, the β-catenin signaling pathway was examined. Spry2 knockdown stabilized β-catenin protein, which might be induced by NEK2 as mentioned in our previous29 and other studies,30 which have documented that β-catenin phosphorylation at Ser33/Ser37 is mediated by NEK2 activity and provides resistance against β-catenin degradation by E3 ligase β-TrCP. We also observed elevated NEK2 and reduced Yap1 protein levels by the suppression of Spry2, whose mechanism warrants further research. Although a significant change in β-catenin protein levels was not detected under an enforced Spry2 expression, increased levels of Spry2 inhibited the activation of β-catenin transcriptional activity and NEK2 kinase activity. Collectively, our results suggest that the suppression of the negative role of Spry2 mediated by c-Cbl E3 ligase elevates EGFR and β-catenin signaling under CUG2 overexpression and maintains activated EGFR and β-catenin signaling, thereby contributing to the induction of cancer stem cell-like phenotypes.
Acknowledgments
We thank Professor Tarun B. Patel (Albany College of Pharmacy and Health Sciences, NY, USA) for providing pHM6-Spry2 vector.
Funding
This study was supported by two-year research grant from Pusan National University, Busan, Republic of Korea.
Disclosure
Natpaphan Yawut and Phatcharaporn Budluang report grants from BK21 Plus, Department of Cogno-Mechatronics Engineering, Pusan National University, Busan 46241, Republic of Korea, during the conduct of the study. The authors declare that they have no other potential conflicts of interest for this work.
References
1. Lee S, Gang J, Jeon SB, et al. Molecular cloning and functional analysis of a novel oncogene, cancer-upregulated gene 2 (CUG2). Biochem Biophys Res Commun. 2007;360(3):633–639.
2. Kaowinn S, Kim J, Lee J, et al. Cancer upregulated gene 2 induces epithelial-mesenchymal transition of human lung cancer cells via TGF-β signaling. Oncotarget. 2017;8(3):5092. doi:10.18632/oncotarget.13867
3. Kaowinn S, Jun SW, Kim CS, et al. Increased EGFR expression induced by a novel oncogene, CUG2, confers resistance to doxorubicin through Stat1-HDAC4 signaling. Cell Oncol. 2017;40(6):549–561. doi:10.1007/s13402-017-0343-7
4. Kaowinn S, Seo EJ, Heo W, et al. Cancer upregulated gene 2 (CUG2), a novel oncogene, promotes stemness-like properties via the NPM1-TGF-β signaling axis. Biochem Biophys Res Commun. 2019;514(4):1278–1284. doi:10.1016/j.bbrc.2019.05.091
5. Kaowinn S, Yawut N, Koh SS, Chung Y-H. Cancer upregulated gene (CUG) 2 elevates YAP1 expression, leading to enhancement of epithelial-mesenchymal transition in human lung cancer cells. Biochem Biophys Res Commun. 2019;511(1):122–128. doi:10.1016/j.bbrc.2019.02.036
6. Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92(2):253–263. doi:10.1016/S0092-8674(00)80919-8
7. Casci T, Vinós J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96(5):655–665. doi:10.1016/S0092-8674(00)80576-0
8. Egan JE, Hall AB, Yatsula BA, Bar-Sagi D. The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins. Proc Natl Acad Sci. 2002;99(9):6041–6046. doi:10.1073/pnas.052090899
9. Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4(11):850–858. doi:10.1038/ncb867
10. Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2(4):294–307. doi:10.1038/35067100
11. Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol. 2004;5(6):441–450. doi:10.1038/nrm1400
12. Sutterlüty H, Mayer C-E, Setinek U, et al. Down-regulation of Sprouty2 in non–small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and-independent mechanisms. Mol Cancer Res. 2007;5(5):509–520. doi:10.1158/1541-7786.MCR-06-0273
13. Lo TL, Yusoff P, Fong CW, et al. The ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res. 2004;64(17):6127–6136. doi:10.1158/0008-5472.CAN-04-1207
14. Fritzsche S, Kenzelmann M, Hoffmann M, et al. Concomitant down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr Relat Cancer. 2006;13(3):839–849. doi:10.1677/erc.1.01190
15. Shaw AT, Meissner A, Dowdle JA, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21(6):694–707. doi:10.1101/gad.1526207
16. Lyle CL, Belghasem M, Chitalia VC. c-Cbl: an important regulator and a target in angiogenesis and tumorigenesis. Cells. 2019;8(5):498. doi:10.3390/cells8050498
17. Haglund K, Schmidt MH, Wong ESM, Guy GR, Dikic I. Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation. EMBO Rep. 2005;6(7):635–641. doi:10.1038/sj.embor.7400453
18. Kales SC, Ryan PE, Lipkowitz S. Cbl exposes its RING finger. Nat Struct Mol Biol. 2012;19(2):131. doi:10.1038/nsmb.2241
19. Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Current Biol. 2003;13(4):308–314. doi:10.1016/S0960-9822(03)00086-1
20. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nat Protoc. 2008;3(6):1101. doi:10.1038/nprot.2008.73
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
22. Park E, Cho I, Srisuttee R, et al. CUG2, a novel oncogene confers reoviral replication through Ras and p38 signaling pathway. Cancer Gene Ther. 2010;17(5):307–314. doi:10.1038/cgt.2009.83
23. Kaowinn S, Kaewpiboon C, Koh SS, Krämer OH, Chung YH. STAT1‑HDAC4 signaling induces epithelial‑mesenchymal transition and sphere formation of cancer cells overexpressing the oncogene, CUG2. Oncol Rep. 2018;40(5):2619–2627.
24. Wong ESM, Lim J, Low BC, Chen Q, Guy GR. Evidence for direct interaction between Sprouty and Cbl. J Biol Chem. 2001;276(8):5866–5875. doi:10.1074/jbc.M006945200
25. Fong CW, Leong HF, Wong ESM, Lim J, Yusoff P, Guy GR. Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J Biol Chem. 2003;278(35):33456–33464. doi:10.1074/jbc.M301317200
26. Wong ESM, Fong CW, Lim J, et al. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. EMBO J. 2002;21(18):4796–4808. doi:10.1093/emboj/cdf493
27. He Y, Ge Y, Jiang M, et al. MiR-592 promotes gastric cancer proliferation, migration, and invasion through the PI3K/AKT and MAPK/ERK signaling pathways by targeting Spry2. Cell Physiol Biochem. 2018;47(4):1465–1481. doi:10.1159/000490839
28. Li J, Yang R, Dong Y, Chen M, Wang Y, Wang G. Knockdown of FOXO3a induces epithelial-mesenchymal transition and promotes metastasis of pancreatic ductal adenocarcinoma by activation of the β-catenin/TCF4 pathway through SPRY2. J Exp Clin Cancer Res. 2019;38(1):38. doi:10.1186/s13046-019-1046-x
29. Dean J, Silva J, McCoy J, et al. Lymphocyte blastogenesis induced by potassium chloride extracts of allogeneic breast carcinoma and lymphoid cells. J Natl Cancer Inst. 1975;54(6):1295–1298. doi:10.1093/jnci/54.6.1295
30. Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–988. doi:10.1016/j.immuni.2006.10.016
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.