Back to Journals » Cancer Management and Research » Volume 12
OTUD4: A Potential Prognosis Biomarker for Multiple Human Cancers
Authors Zhao X, Su X, Cao L, Xie T, Chen Q , Li J, Xu R , Jiang C
Received 4 October 2019
Accepted for publication 14 February 2020
Published 28 February 2020 Volume 2020:12 Pages 1503—1512
DOI https://doi.org/10.2147/CMAR.S233028
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Xiaohui Zhao,1,* Xiaobo Su,1,* Lu Cao,2 Tian Xie,3 Quan Chen,1 Jing Li,1 Rui Xu,4 Chao Jiang1,5
1GMU-GIBH Joint School of Life Sciences, Guangzhou Medical University, Guangzhou 511436, People’s Republic of China; 2Juancheng People’s Hospital, Heze City, Shandong Province 274600, People’s Republic of China; 3Obstetrics and Prenatal Diagnosis Center, Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou 510630, People’s Republic of China; 4Affiliated Cancer Hospital & Institute of Guangzhou Medical University, Guangzhou 510095, People’s Republic of China; 5Department of Cancer Center, People’s Hospital of Baoan District, Shenzhen 518101, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaohui Zhao
GMU-GIBH Joint School of Life Sciences, Guangzhou Medical University, Guangzhou 511436, People’s Republic of China
Tel +86-20-3710-3221
Email [email protected]
Chao Jiang
GMU-GIBH Joint School of Life Sciences, Guangzhou Medical University, Guangzhou 511436, Department of Cancer Center, People’s Hospital of Baoan District, Shenzhen 518101 People’s Republic of China
Tel +86-0755-27788311
Email [email protected]
Background: Deubiquitinase OTU domain containing 4 (OTUD4) is initially identified as a K48-specific deubiquitinase and plays an important role in DNA damage repair signaling transduction. However, the expression level, prognostic role, biological function and mechanism of OTUD4 in multiple human cancers are unclear.
Methods: GEPIA online (http://gepia.cancer-pku.cn/; The Cancer Genome Atlas (TCGA) database) was used to analyze the mRNA expression of OTUD4 in multiple human cancers. Kaplan-Meier plotter (KM plotter) database and TCGA database were used to evaluate the prognostic value of OTUD4 expression in multiple human cancers. MTT, Transwell and 3D culture assays were used to detect the role of OTUD4 in breast, liver and lung cancer cells. The correlation between OTUD4 and apoptosis signaling pathway and AKT signaling pathway was analyzed by Gene set enrichment analysis (GSEA).
Results: OTUD4 mRNA expression is significantly downregulated in multiple human cancer tissues. Survival analysis establishes that the downregulation of OTUD4 predicts poor prognosis in many solid tumors, including breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and ovarian serous cystadenocarcinoma (OV). Furthermore, overexpression of OTUD4 could inhibit tumor cell proliferation, migration and invasion of breast, liver and lung cancer cells through inhibiting the AKT signaling pathway.
Conclusion: This study found that OTUD4 may be a potential predictive factor for several human cancers and a tumor suppressor for breast, liver and lung cancer. The overexpression of OTUD4 restrained proliferation, migration and invasion of human breast, liver and lung cancer cells through promoting cancer cells apoptosis and inhibiting AKT signaling pathway. Notably, our results indicated that OTUD4 could be a useful biomarker for the prognosis of human cancers and a potential molecular target for diagnosis and treatment of breast, liver and lung cancer.
Keywords: OTUD4, prognosis biomarker, proliferation, migration, invasion
Introduction
Cancer, a devastating disease, is the main cause of death in both developed countries and developing countries and constitutes an enormous burden on society. The occurrence of cancer is increasing, and so as the social burden. According to GLOBOCAN estimates, about 18.1 million new cancer cases and 9.6 million deaths occurred in 2018 worldwide.1 The main treatment methods for cancer include traditional methods, surgery, radiation therapy and chemotherapy, and the newly emerging molecular targeted therapy and immunotherapy. The three traditional treatment therapies for cancer have many limitations and evolve slowly over the past decades. For example, surgery often causes significant pain and disfigurement for patients; radiation therapy can only cure local disease; chemotherapy may result in extensive dangerous side-effects and drug resistance. By contrast, molecular targeted therapy and immunotherapy with distinct advantages emerge as promising new modalities for anticancer therapy. Molecular targeted therapy acts on specific tumor targets and can achieve better responses with fewer toxicities. This approach has achieved great success in many human cancers, such as but not limited to: the receptor tyrosine kinase inhibitor (TKI) imatinib in chronic myelogenous leukemia,2 the anti-HER2 monoclonal antibody Herceptin in breast cancer,3 the multi-targets kinase inhibitor sorafenib in liver cancer,4 the third-generation EGFR-TKIs Osimertinib in lung cancer,5 and the humanized monoclonal antibody nimotuzumab in head and neck cancers.6 Cancer immunotherapy offers great hope for a cure by targeting cancer cells more specifically and more robustly than existing traditional treatments. The promise for this method lies in the exquisite specificity of the immune system, which could confer efficacy without toxicity, and the potential for lifelong immunological memory to prevent relapse.2 One of the hottest realms in immunotherapy research is PD-1/PD-L1. In clinical practice, the T cell-directed immune checkpoint inhibitors PD-1/PD-L1 indeed have demonstrated unprecedented efficacy in patients with lung cancer and melanoma. Because of the significant clinical benefits of immunotherapy, increasing clinical trials and extensive basic research on immune checkpoints are gonging on, and the results worth the wait. Theoretically, molecular targeted therapy and immunotherapy may be applied to many types of cancers, so long as cancer has the molecular target or the immune target. It’s a meaningful work to try to find such a molecular target or immune target.
OTUD4, a member of the OTU family, encodes for a protein of 495 amino acids with an OTU domain and functions via their enzymatic deubiquitinating activities.7 Additionally, OTUD4 contains a putative Tudor domain that has been shown to interact with RNA or methylated Histones.8,9 Of the 18 genes with OTU domains in the human genome, 16 genes contain an intact catalytic triad of cysteine, histidine, and aspartate (Ser-His-Asp) to cleave the iso-peptide bond between two ubiquitin units in a poly-ubiquitin chain.10 Human OTUD4, however, only carries a serine (S, Ser) instead of the central cysteine in the catalytic triad (Ser-His-Asp).7 This triad was first identified in the serine proteinases, and the three residues combined in a specific conformation in the active site to hydrolyze the appropriate bond in the substrate.11 This is a ubiquitous set of proteolytic enzymes responsible for a series of physiological reactions, such as the beginning of blood clotting and digestion.12,13 Serine proteinases also play important roles in the tissue destruction associated with arthritis, emphysema, and pancreatitis.14 Previous studies have shown that OTUD4 exerts a DUB-independent function and is mainly used as a scaffold to recruit a multi-DUB complex that directly deubiquitinates the ALKB proteins and then improves the alkylation chemotherapy sensitization of tumors.15 Margolin et al reported homozygous mutations of OTUD4 in patients with ataxia and hypogonadism.16 Silencing OTUD4 in zebrafish embryos leads to defects in the eyes, optic tectum, and cerebellum.16 So far, this is the only report about the deregulation of OTUD4 under pathological conditions. Therefore, we intend to study the relationship between deregulated OTUD4 and multiple human tumors.
In the present study, we reported for the first time that the expression of OTUD4 was downregulated in 11 human cancers. Importantly, our results indicated that the expression of OTUD4 was significantly correlated with the overall survival time of BRCA, ESCA, LIHC, LUAD and OV patients. Furthermore, we also demonstrated that overexpression of OTUD4 significantly reduced the proliferation, migration and invasion of breast, liver and lung cancer cells. Mechanistically, GSEA analysis results indicated that upregulation of OTUD4 may promote cell apoptosis and inhibit AKT signaling pathway. Collectively, these findings suggested that OTUD4 might be a potentially promising biomarker for predicting the prognosis of multiple human cancers and played a tumor suppressor role in human breast, liver and lung cancer.
Materials and Methods
TCGA Dataset Analysis
Gene expression data was downloaded from GEPIA online (http://gepia.cancer-pku.cn/; TCGA database). TCGA Study Abbreviations are shown in Table 1.
 |
Table 1 TCGA Study Abbreviations |
Prognostic Analysis
An online KM plotter database (http://kmplot.com/analysis/) and TCGA database were used to assess the correlation of OTUD4 mRNA expression with cancer patients’ overall survival. Hazard ratio (HR), 95% confidence intervals, and log-rank P were determined and presented on the main plots.
Cell Culture
Breast cancer cells MDA-MB-231, ZR-75-30 and liver cancer cells HepG2, Huh7 were cultured in DMEM medium and lung cancer cells A549, H460 was cultured in RPMI-1640 medium (Gibco, Rockville, MD, USA). The medium was supplemented with 10% FBS, 1% Non-essential amino acids, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco). All cancer cell lines were purchased from ATCC and cultured according to the ATCC protocol.
Plasmids, Transfection and Stable Cell Line Construction
OTUD4 was PCR amplified from cDNA of BEAS-2B cells, subcloned into the BamH I/SpeI sites of pSin-EF2-puro empty vector (EV) and validated by sequencing.17 Plasmids pSin-EF2-OTUD4 was transfected into indicated cells with Lipofectamine-2000 (Invitrogen) according to the manufacturer’s instructions.
RNA Extraction, Reverse Transcription (RT) and Real-Time PCR
Total RNA was extracted using Trizol reagent (Invitrogen). cDNA was synthesized using M-MLV Reverse Transcriptase (Promega). FastStart Universal SYBR Green Master (Roche) was used for quantitative real-time PCR. Primers were as following: OTUD4, forward 5ʹ-AGACCCGAACCAAGCACAT-3ʹ, and reverse 5ʹ-CTGGCTTTTGTTCCGCA-3ʹ; GAPDH, forward 5ʹ-GAAGGTGAAGGTCGGAGTCA-3ʹ, and reverse 5ʹ-TTGAGGTCAATGAAGGGGTC-3ʹ.17 Transcript levels were normalized to the housekeeping gene GAPDH levels. The relative mRNA levels were calculated according to the comparative Ct (ΔΔCt) method, where Ct represents the threshold cycle for each transcript.
Western Blot Analysis
Western blotting was conducted as previously reported.18 The following antibodies were used: anti-OTUD4 (1:500; Millipore) and Anti-α-tubulin (1:5,000; Cell Signaling). Anti-α-tubulin was used as the loading control.
MTT Cell Viability Assay
Cells with a density of 2 × 103 cells per well were inoculated into 96-well plates. At each time point, 100 μL MTT dye (0.5 mg/mL; Sigma-Aldrich Co.) was added to each well and incubated for 4 hrs. Then the culture medium was removed and 100 μL dimethyl sulfoxide (Sigma-Aldrich Co.) was added. The absorbance was determined at 570 nm and at 655 nm. Each experiment was conducted in triplicate.
Cell Migration Assays
Cell migration was measured in Transwell chamber (8 μm; Corning) without Matrigel (BD Biosciences). 2 × 104 indicated cells were suspended in 200 μL serum-free medium and added to the upper chamber, and medium containing 20%FBS was added to the lower chamber. The indicated cells were incubated for 12 or 24 hrs, fixed with paraformaldehyde and then stained with crystal purple. The cells on the bottom surface of the filter membrane were observed and counted under an i200 microscope.
3D Culture Assay
The indicated cells (5 × 103) were suspended in 500 μL medium containing 2% matrigel and then inoculated on the 24-well plate coated with 80 μL Matrigel, and 100 μL 10%FBS medium was added every other day. The cells that formed the 3D spherical structure were photographed every 2 days for 10 days.
Statistical Analysis
All statistical analyses were carried out using the GRAPHPAD PRISM 5 software (GraphPad Software, San Diego, CA, USA). Differences between two groups were analyzed using the two-tailed unpaired Student’s t-test; P < 0.05 was considered statistically significant.
Results
OTUD4 mRNA Expression Is Downregulated in Multiple Human Cancers
To determine the potential effect of OTUD4 in multiple human cancers, we first validated OTUD4 mRNA expression level using the TCGA database, and the results showed that OTUD4 mRNA expression was significantly downregulated in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA) and uterine corpus endometrial carcinoma (UCEC) tissues compared with corresponding normal tissues. Inconsistently, OTUD4 could act as an oncogene in other types of cancer, including pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD), and thymoma (THYM) (Figure 1), for the mRNA expression of OTUD4 in these cancers was markedly upregulated.
OTUD4 Is a Prognostic Biomarker in Several Solid Tumors
To further investigate the prognostic role of OTUD4 in multiple human cancers, we used an online tool (http://kmplot.com) and the TCGA database to explore the relationship between OTUD4 expression and cancer patients’ clinicopathological features. Statistical analysis revealed that OTUD4 expression was significantly positively associated with increased overall survival time of BRCA (HR=0.81; P=0.00013), ESCA (HR=0.41; P=0.027), LIHC (HR=0.54; P=0.023), LUAD (HR=0.47; P=6.6e-10), and OV (HR=0.53; P=0.018, Figure 2) patients. Overall, the results indicated that OTUD4 could act as a tumor suppressor and might be a predictive biomarker for disease outcomes in BRCA, ESCA, LIHC, LUAD and OV patients.
OTUD4 Inhibits Tumor Cells Proliferation and Promotes Tumor Cells Apoptosis
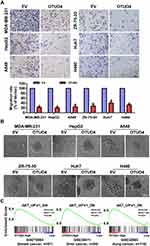
In view of the low expression of OTUD4 in multiple human cancers, we studied the role of OTUD4 in the proliferation of breast, liver, and lung cancer cells. MDA-MB-231, ZR-75-30, HepG2, Huh7, A549 and H460 cells were transfected with OTUD4 or EV by Lipo2000. As shown in Figure 3A, Real Time PCR and Western blot analysis confirmed that OTUD4 mRNA and protein levels were significantly elevated after transfection. MTT assay indicated that the proliferation abilities of these cancer cell lines were dramatically inhibited after OTUD4 transfection (Figure 3B). To further study the reasons why OTUD4 affects the proliferation of tumor cells, Gene set enrichment analysis (GSEA) was used to analyze the relationship between the expression of OTUD4 and cell cycle and apoptosis of multiple human cancer cells. GSEA revealed that OTUD4 expression was positively associated with apoptosis signatures (HALLMARK_APOPTOSIS) in a publicly available GEO database of breast (GEO42568), liver (GSE26566) and lung cancer (GSE3141) (Figure 3C). In addition, GSEA results showed that the expression of OTUD4 does not regulate the operation of cell cycle. These results indicated that OTUD4 inhibits the proliferation of multiple cancer cells through promoting the apoptosis of those cancer cells.
OTUD4 Inhibits Migration and Invasion Through Inhibiting AKT Signaling Pathway
To further determine the migratory ability of breast, liver and lung cancer cells, Transwell without matrigel assay was performed, and the result was shown in Figure 4A. The migration rate of MDA-MB-231, ZR-75-30, HepG2, Huh7, A549 and H460 cells transfected with OTUD4 was drastically lower than that of EV cells. In addition, in 3D culture assay, OTUD4-transduced cancer cells grew into less invasive processes than EV cells (Figure 4B). In summary, these findings suggested that OTUD4 played a vital role in the migration and invasion of breast, liver and lung cancer cells.
To explore the mechanism of OTUD4-mediated proliferation, migration and invasion in breast, liver and lung cancer cells, we next studied the relationship between OTUD4 expression level, and the genes regulated by various signaling signatures using GSEA of GEO datasets. GSEA results showed that OTUD4 expression is positively correlated with the AKT-downregulated gene signature (AKT_UP. V1_DN) in a publicly available GEO database of breast (GEO70884), liver (GSE36411) and lung cancer (GSE32865) (Figure 4C), indicating that OTUD4 may inhibiting the AKT signaling pathway.
Discussion
OTUD4 is originally identified as a deubiquitinating enzyme with a preference for K48-linked chains. Liuyu et al demonstrated that constant RNA virus infection induces K48-linked ubiquitination and degradation of the adapter protein MAVS and the upregulation of OTUD4. OTUD4 interacts with MAVS and removes the K48-linked polyubiquitin chain from MAVS to prevent excessive degradation of MAVS, thus maintaining the antiviral response to a certain level and limiting the replication of invading viruses by inhibiting NF-κB signaling.19 Yet, Zhao et al reported that OTUD4 directly controls MyD88 K63 deubiquitinating enzyme activity to suppress IL-1β-dependent NF-κB signaling, which is not only dependent on the phosphorylation of OTUD4 near its catalytic domain but also dependent on a putative ubiquitin-interacting motif adjacent to the OTU domain.20 According to another published study, deregulated OTUD4 indeed affects the levels of ubiquitinated XPC in human cells, supporting the hypothesis that the OTUD4 deubiquitinating enzyme participates in the XPC recycling by cleaving the ubiquitin moiety.21 Wu et al showed that overexpression of OTUD4 could damage the homologous recombination repair of DNA double-strand breaks impairs, enhances cell cycle arrest and increases cell death induced by ionizing radiation.17
It is worth mentioned that the involvement of ubiquitinating enzyme in the regulation of cell process sometimes belongs to non-protein degradation, sometimes to protein degradation, and sometimes to the synergistic effect of these two functions.22,23 In this study, we systematically confirmed for the first time that OTUD4 was significantly downregulated in 12 human solid tumor tissues compared with normal tissues. The analysis of Kaplan–Meier Plotter (http://kmplot.com) and TCGA datasets demonstrated that the expression of OTUD4 was positively correlated with the prognosis of BRCA, ESCA, LIHC, LUAD and OV patients. The overall survival time of patients with OTUD4 high expression was significantly longer than that of patients with OTUD4 low expression. These results suggest that OTUD4 has a tumor-suppressing role in multiple human cancers. Furthermore, our results showed that the overexpression of OTUD4 inhibits the proliferation, migration and invasion of breast, liver, and lung cancer cells through inhibiting the AKT signaling pathway. These results suggested that OTUD4 may be a therapeutic target for multiple human cancers.
OTUD4 plays a key role in maintaining the stability of alkylation repair enzymes by promoting the stability of ALKBH3.15 ALKBH3 does not depend on the deubiquitinating enzyme activity of OTUD4, but on the deubiquitinating enzyme activity of its interactors, deubiquitylating enzyme USP7 and USP9X.15 Recent studies have shown that USP7 binds to nuclear ubiquitin PTEN, induce PTEN deubiquitination and nuclear translocation, thus promoting tumor progression.24 Some studies have shown that USP7 also promotes the deubitization and degradation of p53.25 These results suggest that USP7 is a tumorigenic event that promotes the development of cancer. USP9X has the function of stabilizing the expression of MCL1, an important gene that can stabilize progenitor cells and stem cells and maintain the stability of cancer cells.26 Some studies have shown that patients with overexpression of USP9X have a poor prognosis. These results show that USP9X is a prognostic marker for cancer patients. It may also become a new target for cancer therapy.27 Deubiquitination may be a mechanism by which cancer cells maintain stability.
The overall survival time of pancreatic adenocarcinoma (PAAD) patients with low expression of OTUD4 was significantly longer than that of patients with high expression of OTUD4. OTUD4 seems to be an oncogene in pancreatic adenocarcinoma (PAAD). In the human body, the mechanism of gene expression regulation is extremely complicated. In addition to positive feedback regulation, there is also negative feedback regulation.28 Under the condition of negative feedback regulation, it usually involves complex interacting networks in regulating gene expression, tumorigenesis and development. In different types of cancers, even the same gene may play different roles.29 Thus, the expression and function of OTUD4 varies in different normal and tumor tissues and may be cell- and tumor-type specific. The role of OTUD4 is variable, so it should be examined separately in each type of tumors to determine whether it plays a role as a tumor promoter or suppressor. In general, deeply further studies are needed to detail explain the regulatory mechanism of OTUD4 in different cancers. We plan to conduct more scientific work to clarify the functions and regulatory mechanisms of OTUD4 in PAAD in the future.
Overall, in this study, our results suggest that OTUD4 downregulation is associated with poor prognosis in multiple types of human cancers. Overexpression of OTUD4 inhibits proliferation, migration, and invasion of breast, liver and lung cancer cells. Our results suggest that OTUD4 has a tumor-suppressing function and maybe a potential prognostic factor for multiple solid tumors.
Conclusion
This is the first report to systematically assess the potential role of OTUD4 as a clinically independent prognostic factor for disease progression, prognosis, and survival in BRCA, ESCA, LIHC, LUAD and OV patients. In addition, our study demonstrated that overexpression of OTUD4 could inhibit the proliferation, migration and invasion of breast, liver and lung cancer cells through promoting cell apoptosis and inhibiting the AKT signaling pathway. Therefore, it is worthwhile to further investigate the diagnostic and therapeutic value of OTUD4 in multiple human cancers.
Funding
This work was supported by Guangzhou Medical University High-level University Academic Key Cultivation Program (Grant No. B185004137).
Disclosure
The authors report no conflicts of interest in this work.
References
1. BrayF, FerlayJ, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA CANCER J CLIN. 2018;68394–424
2. Helmy KY, Patel SA, Nahas GR, Rameshwar P. Cancer immunotherapy: accomplishments to date and future promise. Ther Deliv. 2013;4(10):1307–1320. doi:10.4155/tde.13.88
3. Ahmed S, Sami A, Xiang J. HER2-directed therapy: current treatment options for HER2-positive breast cancer. Breast Cancer. 2015;22(2):101–116. doi:10.1007/s12282-015-0587-x
4. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15(10):599–616. doi:10.1038/s41571-018-0073-4
5. Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. 2019;4:61. doi:10.1038/s41392-019-0099-9
6. Xu MJ, Johnson DE, Grandis JR. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 2017;36(3):463–473. doi:10.1007/s10555-017-9687-8
7. Louis M, Hofmann K, Broemer M. Evolutionary loss of activity in De-Ubiquitylating enzymes of the OTU family. PLoS One. 2015;10(11):e0143227. doi:10.1371/journal.pone.0143227
8. Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22(2):51–52. doi:10.1016/S0968-0004(96)30049-2
9. Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280(31):28476–28483. doi:10.1074/jbc.M414328200
10. Mevissen TE, Hospenthal MK, Geurink PP, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154(1):169–184. doi:10.1016/j.cell.2013.05.046
11. Blow DM, Birktoft JJ, Hartley BS. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969;221(5178):337–340. doi:10.1038/221337a0
12. Mann KG, Jenny RJ, Krishnaswamy S. Cofactor proteins in the assembly and expression of blood clotting enzyme complexes. Annu Rev Biochem. 1988;57:915–956. doi:10.1146/annurev.bi.57.070188.004411
13. Sigler PB, Blow DM, Matthews BW, Henderson R. Structure of crystalline -chymotrypsin. II. A preliminary report including a hypothesis for the activation mechanism. J Mol Biol. 1968;35(1):143–164. doi:10.1016/S0022-2836(68)80043-9
14. Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci. 1996;5(6):1001–1013. doi:10.1002/pro.5560050603
15. Zhao Y, Majid MC, Soll JM, Brickner JR, Dango S, Mosammaparast N. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J. 2015;34(12):1687–1703. doi:10.15252/embj.201490497
16. Margolin DH, Kousi M, Chan YM, et al. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N Engl J Med. 2013;368(21):1992–2003. doi:10.1056/NEJMoa1215993
17. Wu Z, Qiu M, Guo Y, et al. OTU deubiquitinase 4 is silenced and radiosensitizes non-small cell lung cancer cells via inhibiting DNA repair. Cancer Cell Int. 2019;19:99. doi:10.1186/s12935-019-0816-z
18. Wu Z, Zhao J, Qiu M, et al. CRISPR/Cas9 mediated GFP knock-in at the MAP1LC3B locus in 293FT cells is better for bona fide monitoring cellular autophagy. Biotechnol J. 2018;13(11):e1700674. doi:10.1002/biot.v13.11
19. Liuyu T, Yu K, Ye L, et al. Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 2019;29(1):67–79. doi:10.1038/s41422-018-0107-6
20. Zhao Y, Mudge MC, Soll JM, et al. OTUD4 is a phospho-activated K63 deubiquitinase that Regulates MyD88-dependent signaling. Mol Cell. 2018;69(3):505–516 e505. doi:10.1016/j.molcel.2018.01.009
21. Lubin A, Zhang L, Chen H, White VM, Gong F. A human XPC protein interactome–a resource. Int J Mol Sci. 2013;15(1):141–158. doi:10.3390/ijms15010141
22. Woelk T, Oldrini B, Maspero E, et al. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8(11):1246–1254. doi:10.1038/ncb1484
23. Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi:10.1016/j.molcel.2009.01.014
24. Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys. 2011;60(1–2):61–68. doi:10.1007/s12013-011-9185-5
25. Sheng Y, Saridakis V, Sarkari F, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13(3):285–291. doi:10.1038/nsmb1067
26. Trivigno D, Essmann F, Huber SM, Rudner J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia. 2012;14(10):893–904. doi:10.1593/neo.12598
27. Schwickart M, Huang X, Lill JR, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–107. doi:10.1038/nature08646
28. Ferrell JE
29. Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: tumor promoter or suppressor? Biochim Biophys Acta. 2011;1816(1):80–88. doi:10.1016/j.bbcan.2011.04.004
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.