Back to Journals » Journal of Inflammation Research » Volume 15
Oral-Intestinal Microbiota in Colorectal Cancer: Inflammation and Immunosuppression
Authors Mo S, Ru H, Huang M, Cheng L, Mo X, Yan L
Received 14 October 2021
Accepted for publication 7 January 2022
Published 4 February 2022 Volume 2022:15 Pages 747—759
DOI https://doi.org/10.2147/JIR.S344321
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Sisi Mo,1– 3,* Haiming Ru,1– 3,* Maosen Huang,1– 3 Linyao Cheng,1– 3 Xianwei Mo,1– 3 Linhai Yan1– 3
1Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China; 2Guangxi Clinical Research Center for Colorectal Cancer, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China; 3Guangxi Key Laboratory of Colorectal Cancer Prevention and Treatment, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Linhai Yan
Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital, Hedi Road No. 71, Nanning, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China
, Tel +86 139 78839969
, Email [email protected]
Abstract: It is widely recognized that microbial disorders are involved in the pathogenesis of many malignant tumors. The oral and intestinal tract are two of the overriding microbial habitats in the human body. Although they are anatomically and physiologically continuous, belonging to the openings at both ends of the digestive tract, the oral and intestinal microbiome do not cross talk with each other due to a variety of reasons, including intestinal microbial colonization resistance and chemical barriers in the upper digestive tract. However, this balance can be upset in certain circumstances, such as disruption of colonization resistance of gut microbes, intestinal inflammation, and disruption of the digestive tract chemical barrier. Evidence is now accruing to suggest that the oral microbiome can colonize the gut, leading to dysregulation of the gut microbes. Furthermore, the oral-gut microbes create an intestinal inflammatory and immunosuppressive microenvironment conducive to tumorigenesis and progression of colorectal cancer (CRC). Here, we review the oral to intestinal microbial transmission and the inflammatory and immunosuppressive microenvironment, induced by oral-gut axis microbes in the gut. A superior comprehension of the contribution of the oral-intestinal microbes to CRC provides new insights into the prevention and treatment of CRC in the future.
Keywords: oral-intestinal microbiota, colorectal cancer, inflammatory microenvironment, immunosuppressive microenvironment
Introduction
Symbiotic microbes, numbering in their millions of billions, colonize the human body, including the skin, oral, and digestive tract.1 The oral and gastrointestinal tract are two of the largest microbial habitats2 and play significant roles in microbiome related diseases.3,4 Although the mouth and gut are connected by the digestive tract, communication between the two microbial ecosystems does not often lead to disease. This is as a consequence of the oral-intestinal barrier which exists, including gastrointestinal acid5 and bile acid6 chemical barriers in the digestive tract, intestinal microbial colonization resistance,7 and host pattern recognition receptors.8 But there are conditions in which the barrier is broken down, such as when particular drugs as utilised9,10 or when the gut is exposed to chronic inflammation.11 At this point, the oral microbiome spreads to the gut and has the opportunity to colonize as pathogens, thus reshaping the intestinal microbiota system. This process is often associated with disease states.12
Colorectal cancer (CRC) is a common malignant disease, representing the third and second highest incidence among cancers in men and women respectively, and is the second highest cause of cancer-related death globally.13 Most CRCs are sporadic and only a few cases are inherited or associated with specific susceptible diseases, for instance, inflammatory bowel disease (IBD).14 CRC is a multi-factor process, involving genetic, epigenetic, and environmental factors.15,16 Recently, the contribution of microbial disorders to CRC has become more prominent. Accumulated evidence has indicated that microbial dysregulation plays a considerable role in the tumorigenesis of CRC.17,18
The most widely recognized microbe association with cancer is Helicobacter pylori infection and its interactions with gastric cancer.19 In addition, research has also found that oral pathogens have a place in the occurrence and development of CRC,20 including Fusobacterium nucleatum (F. nucleatum)21 and Porphyromonas gingivalis (P. gingivalis),22 which are the most widely implicated to date. Most of these pathogens are common pathogenic bacteria observed in periodontitis, causing an inflammatory environment in periodontal tissue and regulating immune responses.23,24 However, the influence of oral pathogens on the host intestinal environment, following transmission to the lower digestive tract, remains to be explored. Present data suggests that microbes communicate between organs, for example in the gut-brain axis25 and the gut-liver axis,26 which often represent pathological processes. Therefore, attention should be given towards understanding the crosstalk of microorganisms between various organs.
In this review, the ways in which oral microbes spread to the gut and colonize as opportunistic pathogens are focused on, and their mechanisms in tumorigenesis and the progression of CRC are summarized.
Oral and Intestinal Microbiome: Communication and Insulation
Oral and Intestinal Districts Communicate Through the Digestive Tract
The human digestive system is made up of digestive tubes, the tubes that run from the mouth to the anus, and the digestive glands. As a result, oral and intestinal mucosa are biologically connected, and oral microbes may migrate to the lower digestive tract.27 Saliva and food, secreted or ingested by the oral cavity, pass through the gut, resulting in a chemical connection between the oral and intestinal tracts. However, the oral and gut microbiota are two communities with very distinct characteristics.
Oral Microbial Community
The oral system is open to the environment, and it interacts with microbes with every breath and diet. About 760 species of microbes inhabit the oral tract, colonizing in various habitats, such as the teeth, tongue, buccal mucosa, and gums, depending on the chemical constitution, morphology, and stability of the oral cavity.28 The Human Microbiome Project (HMP) collected samples from nine different habitats in the oral system, covering more than half of the total oral habitats, including saliva; buccal mucosa (cheek), keratinized gingiva (gums), palate, tonsils, throat, and tongue soft tissues; and supra- and subgingival dental plaque (tooth biofilm above and below the gum) to map the distribution of oral microbiome in a healthy population. The results showed that the microbial taxa of various habitats in the oral cavity were highly personalized. Although Streptococcuspredominates most habitats, the chief inhabitant in buccal mucosa was found to be Haemophilus, with the supragingival plaque populated by Actinomyces, whereas Prevotella was the most abundant bacteria in the subgingival plaque.29 The highly personalized oral microbiome is also reflected in the difference between genus and species. The oral cavity is routinely composed of the same genera, mainly Actinomyces, Fusobacterium, Neisseria, Veillonella, and Rothia. However, their individual species show strong differences in habitat distribution.30 Oral microbiome in the healthy state is related to time, age, diet and environment.31 Numerous oral microbes interact with each other to help the body resist adverse external stimuli. However, imbalances in the oral microbiome can lead to oral diseases such as dental caries and periodontitis,32 and systemic diseases such as IBD33 and Alzheimer’s disease.4 Therefore, the oral microbiome is a key link in balancing microbiome homeostasis and health.
Intestinal Microbial Community
The gut has been described a secret garden of microbes, home to 10 trillion different symbiotes, including 50 bacterial phyla and about 100–1000 bacterial species. The anaerobic environment of the gut allows seven phyla to thrive: Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Proteobacteria, Verrucomicrobia and Cyanobacteria. Among them, Bacteroidetes and Firmicutes were found to dominant more than 90% of the population.34 The ratio of Firmicutes and Bacteroidetes changes under physiological and pathological conditions and is considered a predictor of health and disease.35,36 In addition to phylum classification, the human intestinal microbiome is divided into three enterotypes according to species and functional modules. Enterotype 1 is enriched in Bacteroides with a very broad glycolysis potential. Enterotype 2 dominant by Prevotella with its ability to degrade mucin glycoproteins. Enterotype 3 is the most frequent of the three and is enriched in Ruminococcus with mucin degrading activities and membrane transportation of sugars.37 Gut microbes are influenced by age, diet, and environment, and participate in the hosts metabolic, immune, and neural activities.38–41 The intestinal microbes are resistant to colonization by foreign microorganisms through direct and indirect mechanisms, namely colonization resistance (CR) to pathogens.42 The direct mechanism is characterized by the ability of the symbiotic microbiome to restrict the colonization of exogenous microorganisms and hold back from pathological overgrowth of the original inhabitants strictly via its own factors without needing to resort to any host interaction, including competition for nutritional and physical space,43 Type VI pituitary systems,9 bacteriophages and inhibitory metabolites as well as bactericidal molecules produced by gut microbiota,7 such as bacteriocins, short chain fatty acids (SCFAs), and secondary bile acids. Indirect mechanisms are characterized by the symbiotic microbiota’s dependence on factors, which originate in the hosts resistance to foreign pathogens, such as the epithelial barrier, innate immune defense system and an adaptive immune mechanism.44 Their specific contributions to CR have been widely described7,9,42,45 and will not be repeated here.
Oral Bacteria Associated with CRC
Oral Bacteria are Enriched in CRC
Several recent studies have demonstrated the involvement of oral bacteria in tumorigenesis and development of CRC by metagenomic or 16S rRNA sequencing. Yang et al46 summarized the profiles of gut microbes in fecal samples from healthy controls and patients suffering from CRC. Compared with healthy controls, the balance of gut bacteria in CRC patients was disrupted and their abundance was reduced. Intestinal microbial diversity has also been shown to be strongly influenced by tumor load. At the generic level, Parvimonas, Fusobacterium and Porphyromonas, which are commonly found in the oral cavity, were enriched in CRC patients. At the species level, Dai et al47 characterized fecal microbial differences between healthy controls and CRC patients from different countries. Metagenomic sequencing showed that seven CRC-enriched species, including four oral bacteria: Porphyromonas asaccharolytica, F. nucleatum, Prevotella intermedia, and Parvimonas micra (P. micra), were able to distinguish healthy controls from CRC patients. Interestingly, bacteria also form mutually beneficial networks. Moreover, F. nucleatum was located in the center of this network, that is, oral bacteria may play a crucial role in the network. Strong evidence for this hypothesis was that F. nucleatum contributed towards the entry and colonization of other bacteria into biofilms, a potential source of inflammation and tumorigenesis in CRC.48 The intestinal mucosa, as the interface of bacterial biofilm, has also shown evidence of oral bacteria. Russo et al49 revealed that the composition and abundance of microbes varied in different sites within CRC patients. This phenomenon has been clearly seen in cancer tissues and stool samples from CRC patients. Proteobacteria and Fusobacteria were abundant in cancer tissues, while Firmicutes and Fusobacteria were found to be more abundant in stool samples. The study also confirmed the presence of F. nucleatum in saliva, stool and cancer tissue samples from CRC patients. In addition, abundance of F. nucleatum in cancer tissues was higher than that observed in stool samples. This means that bacteria from the oral system can spread and colonize the gut. Another study detected identical F. nucleatum strains in cancer tissue and saliva samples from CRC patients, strengthening the hypothesis that oral bacteria can colonize the intestinal tract and contribute towards CRC tumorigenesis.50 In order to explore whether there were differences in intestinal microbes between patients with differing stages of CRC, a cohort study that included early and late CRC found that P. micra, Peptostreptococcus stomatis, Solobacterium moorei, Gemella morbillorum and F. nucleatum had increased abundance in both early and late stage, while abundance of Actinomyces odontolyticus increased only at an early stage.51 In addition, gut microbiota in CRC patients showed specificity in proximal and distal carcinoma tissues. Distal tumors have been characterized by higher abundances of Alistipes, Akkermansia, Halomonas and Shewanella, whereas Faecalibacterium, Blautia and Clostridium were more abundant in proximal tumors. The above changes regarding composition and abundance of oral bacteria in CRC and healthy controls are summarized in Table 1.
 |
Table 1 Difference in the Abundance of Oral-Intestinal Microbes in CRC vs Healthy Controls |
These studies support the hypothesis that oral bacteria spread down the digestive tract and colonize the gut. The abundance of oral bacteria in stool samples was lower than in cancerous tissue, indicating that the microbes in stool were diluted. This may be related to the accumulation of oral bacteria in the intestinal mucosa to form biofilms and the addition of other bacteria.
Potential Protective Effects of Oral Bacteria on Gut
Although the contribution of oral bacteria to tumorigenicity of CRC was reported in the previous study,20 some oral bacteria are still beneficial to health. Streptococcus salivarius, as one of the earliest inhabitants of the oral system, it helps to balance immune homeostasis and regulate inflammatory response. A study has shown that Streptococcus salivarius not only inhibits the activation of NF-κB in intestinal epithelial cells and prevents inflammatory responses, but also inhibits the inflammatory response in mouse models of colitis.52 The beneficial mediators were derived from the metabolic products of Streptococcus salivarius, because these reactions were not observed in heat-inactivated bacteria. Oral microbes have been associated with inflammatory responses in IBD patients, suggesting that oral microbes are involved in intestinal immune and inflammatory processes. A previous study observed that the abundance of Streptococcus, Rothia, Neisseria, Haemophilus, and Gemella, were decreased in IBD patients compared to healthy controls, and were positively correlated with immunological biomarkers in IBD patients, such as lysozyme, and negatively correlated with IL-1β and IL-8 levels. Lysozyme is an antibacterial protein and plays an important role in the host defense system.53
Translocation of Microbes from Oral to Gut
How Do Oral Microbes Travel to the Gut?
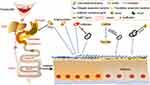
It is worth noting that traces of classic oral microbes have been found in gastrointestinal pathological conditions. For example, the intestinal mucosa from patients who suffered from IBD was significantly enriched with Porphyromonas, Prevotella and Gemella.33 This suggests that the invasion and colonization of oral pathogens contribute to intestinal pathologic changes, when mucosal homeostasis is disrupted. There are two main ways which the oral microbiome is thought to be transmitted to the gut. One is daily chewing and swallowing, given that people swallow about 1.5 liters of saliva daily, accompanied by millions of oral microbes. The researchers suggested that most of these microbes die as they cross the gastrointestinal barrier.2 In ApcMin/+ mouse models, oral F. nucleatum increased colonic tumor load and could be isolated from tumors.54 Although traces of oral microbes can also be found in stool samples from healthy people. However, people with CRC have more oral and intestinal transmission than healthy people.55,56 This suggests that in some cases, oral microbes colonize the gut and become opportunistic pathogens. The other way is through the hematogenous or lymphatic pathway. Recent research confirmed that the oral microbiome can enter the bloodstream directly during dental activities such as brushing and tooth extraction, as well as daily activities such as chewing.57 During chronic periodontitis, the periodontal blood vessels proliferate and dilate, and the gingival epithelium is more permeable in an inflammatory environment, allowing microbe to infiltrate and spread to external areas of the mouth, including joints and the colon.58 The circulatory system seems as the most effective way for F. nucleatum to reach CRC during the brief physiological bacteremia of the oral.59 A study can be used as evidence of a hematogenous or lymphatic pathway. Fusobacterium can be detected in liver metastases of CRC and is consistent with the strain found in the primary tumor.60
How Do Oral Microbes Colonize the Gut?
Colonization Resistance Disruption in Gut Microbiota
As mentioned above, the gut microbes provide CR against foreign microbial invasion. Certain medications, such as antipsychotics and PPIs,7 affect the intestinal microbial ecosystem and threaten CR, of which antibiotics represent the most widespread concern. The use of antibiotic leads to a temporary or severe loss of intestinal microbial diversity and species, with reduced microbial concentrations present in people using antibiotics.9 This has been characterized by a decrease in anaerobic bacteria (Lactobacillus, Clostridium, Bacteroides, and Bifidobacterium), which are replaced by increases in Enterococcus and Enterobacter (Klebsiella, Enterobacter, and Citrobacter), resulting in decreased numbers of SCFAs in the gut.61 Antibiotic treatment causes intestinal macrophages to become overactive, promoting persistent T-cell-mediated dysfunction.62 These changes increase the susceptibility of the gut to a range of pathogens, including oral pathogens. Klebsiella infection is the most representative example. Klebsiella encodes an antibiotic resistance gene, and mouse models treated with ampicillin or tylosin resulted in Klebsiella colonization in the intestinal tract, while mouse models not treated with antibiotics were resistant to colonization within their intestinal tracts. Klebsiella has been found to be associated with inflammation-induced CRC. Moreover, epithelial tumor necrosis factor-α (TNF-α) mRNA expression and inflammatory mediators were observed in germ-free (GF) IL10−/− + Klebsiella pneumoniae 2H7 mouse model in comparison with other mouse models (GF WT+ Escherichia coli (E. coli) or GF IL10−/− + E. coli).63 In the case of PPIs, the effect on gut microbiome composition is thought to be the result of a combination of the two mechanisms. One is to indirectly change the PH of the gastrointestinal tract, creating an environment suitable for oral bacteria to pass through and grow. The other is direct inhibition of certain symbiotic gut bacteria that produce SCFAs, such as Lachnospiraceae and Ruminococcaceae,64 leading to weakened CR of the gut microbiota. In addition to alterations in intestinal microbiota composition and abundance, which is often reported, we demonstrated that in a mouse stress model PPIs increases intestinal permeability by altering factors affecting intestinal mucosal barrier, such as corticotropin-releasing hormone -mast cell- vasoactive intestinal peptide axis and tight junction protein genes.65 Antipsychotics can also affect CR in two ways: one is the recently discovered potential antibacterial effect of antipsychotics,66 and the other is increasing intestinal permeability in the distal ileum of rats.67 This leads to accidental injury of symbiotic intestinal bacteria, which adversely affects CR.
Intestinal Inflammation
A key driver of intestinal environmental changes is the host’s inflammatory response. Human intestinal inflammation is related to the imbalance of the microbiome, which is distinguished by its decreased microbial diversity, reduced abundance of obligate anaerobic bacteria, and expansion of facultative anaerobic bacteria, mainly Enterobacteriaceae members of Proteobacteria.68,69 When the host gut is inflamed, the pattern recognition receptors involved in intestinal immunity are damaged. Especially in IBD, mutation of the NOD2 gene resulted in the reduction of antimicrobial peptides produced by Paneth cells. NOD2 mutants also failed to recruit ATG16L1, resulting in impaired autophagy in epithelial cells.70 Maladjustment of these mechanisms results in reduced bacterial clearance. In addition, oral pathogens have their own nutritional competitive advantage in an inflamed intestinal tract. Enterobacteria, including the portion translocated from the oral mucosa, can utilize diet-derived L-serine catabolic metabolism rather than undertaking monosaccharide metabolism (the way Enterobacteria obtain nutrients under normal intestinal conditions), giving them with the advantage of competition growth compared to the indigenous microbes.71 This is determined by the metabolic profile of the bowel reprogrammed during inflammation. Iron (Fe) is an essential nutrient for Bacteroidetes, the dominant microbes in the gut. However, in the inflamed gut, the host limits free Fe concentrations in the blood via a variety of mechanisms.72 The P. gingivalis Hmu heme acquisition system is dominated by HmuY and competes with other members of HmuY-like proteins to increase P. gingivalis virulence and its ability to also cause dysbiosis in the gut microbiome. In addition, HmuY was not digested when in a suitable medium for Bacteroides, indicating that HmuY was resistant to proteolytic activity of various proteases, therefore conducive to P. gingivalis survival in the intestinal tract.71
Resistance to Chemical Barriers in the Gastrointestinal Tract
As oral pathogens travel down the digestive tract, they pass through two chemical barriers: gastric and bile acids. Gastric acid is one of the key components of the chemical barrier, creating a harsh environment for oral pathogens. In the context of achlorhydria, microbial diversity were decreased and the abundance of oral microorganisms were significantly increased, including P. micra, Peptostreptococcus stomatis, F. nucleatum and Gemella.73 Jackson et al64 demonstrated that in PPIs users there was a weakened role of the gastric acid barrier against foreign microbes, allowing oral microorganisms to develop down in the gastrointestinal tract and colonize. Bile is another barrier to colonization of the intestinal tract by pathogenic microorganisms.74 Erik J Boll discovered that Klebsiella pneumoniae C3091 was the dominant colonizer of the gastrointestinal tract and successfully identified the genes that encode proteins that facilitate colonization of Klebsiella pneumoniae, which did not appear in the E. coli EPI100 strain. These were the RecA recombinase; UDP-galactose-4-epimerase (GalE) and galactose-1-phosphate uridylyltransferase (GalT) from the galactose operon and the ArcA response regulator. Additionally, Klebsiella pneumoniae-derived galET genes aided E. coli colonization by reducing susceptibility to bile salts.75 These studies indicated that certain oral pathogens have evolved mechanisms to resist both gastric and bile acid barriers in order to survive harsh gastrointestinal environment (Figure 1).
Oral-Gut Axis Microbiome Induces an Inflammatory and Immunosuppression Environment in the Gut
Oral-Gut Axis Microbiome Promotes CRC by Activating Inflammasome and NF-κB Cascade Pathways
Accumulating evidence supports the association between chronic inflammation and CRC, and a persistent inflammatory environment promotes the development of CRC.76 The colonization of oral pathogens in the gut can shape the intestinal inflammatory microenvironment, which is facilitated the occurrence and progression of CRC.
Oral microbes were derived from healthy (HOM) and periodontitis (LOM) mice and reconstructed in GF Il10−/− mice. Significantly increased colonic inflammation was observed in LOM colonized mice. Subsequently, the microbial communities from the LOM and HOM models were cultured, with results showing that Enterobacteriaceae were predominant in LOM. Notably, the amassing of TH17 and TH1 in sLOM-colonized mice was observed by mixing the dominant bacteria of LOM and HOM into sLOM and sHOM and reconstituting them in GF Il10−/− mice. Moreover, IL-1β secreting cells were clearly increased in sLOM-colonized mice, while IL-1β secretion was not induced in sHOM-colonized mice.27 TH17 and TH1 are the polarization modes of CD4+T cells and are members of the pro-inflammatory effector Th cells.77 IL-1β makes a critical difference in oral pathogen-driven colitis.53,78
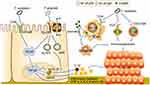
Several classic oral pathogens have been found to induce inflammation-related CRC. Induction of inflammatory microenvironment in CRC involves activation of inflammatory corpuscles and NF-κB.79 F. nucleatum has been shown to secrete outer membrane vesicles in the human colonoid (organoid) monolayers, activate the NF-κB pathway through TLR4, and promote the production of pro-inflammatory cytokines.80 The NF-κB cascade is a central link in the communication within host-microbial interactions during the occurrence and development of CRC. It induces an inflammatory microenvironment by promoting the production of pro-inflammatory cytokines, containing TNF-α, IL-1β, IL-6 and IL-8.81 Increased production of IL-8, IL-1β, and TNF-α was indeed observed in F. nucleatum infected mouse models.82 Additionally, F. nucleatum regulates the expression levels of cancer carcinogenic target MiR-21 through the TLR4/MyD88/NF-κB pathway,83 and then regulates the secretion of TNF-α, IL-6, IL-17A and IL-21.84
The inflammasome is another key factor in inflammation-related CRC, which mainly contributes to the occurrence of CRC by regulating inflammatory cytokines.85 P. gingivalis secretion of nucleotide-diphosphate-kinase enzyme activates P2X7 receptor of colonic epithelium, resulting in opening of P2X7 receptor-mediated ion channels. This opened the pan nexin 1 hole in the colonic epithelial cell membrane, and allowing microbial molecules to enter the cytoplasm, which contributed towards the survival and persistence of intracellular bacteria.86 Subsequently, P. gingivalis activated NF-κB through TLR to increase pro-IL-1β transcription levels, which recruited tumor-infiltrating myeloid cells, and activated the NLPR3 and AIM2 inflammasome to promote maturation of pro-IL-1β into IL-1β, causing a pro-inflammatory microenvironment conducive to CRC progression.87,88 Wang et al infected the myeloid cell line THP1 with P. gingivalis, and within the supernatant increased levels and secretion of TNF-α, IL-6, and IL-1β were detected.87
Oral-Gut Axis Microbiome Promotes CRC by Creating Immunosuppressive Microenvironment
Toll-like receptors (TLRs) are pivotal in the innate immune system of the host by recognizing the pathogen-associated molecular patterns of microorganisms.89 Different subtypes of TLRs were expressed in different types of intestinal cells, including intestinal epithelial cells, immune cells, and parenchymal non-immune cells. Among them, TLR4, TLR5, TLR9, TLR2 and its heterodimerizing partners, TLR1 and TLR6 specialize in identifying bacteria.90 F. nucleatum has an immunosuppressant effect by promoting the M2 polarization of macrophages (M2-Mφ) both in vivo and in vitro through TLR4-dependent mechanism. It is worth mentioning that the F. nucleatum strain derived from enterogenous (F01) induced a higher level of M2-Mφ than that from the oral cavity (ATCC10953). This suggests that oral F. nucleatum may have evolved to a higher level of virulence after intestinal colonization. This polarization activation involves the cascade signaling pathway of IL-6/P-STAT3/C-MYC91 and TLR4/NF-κB/S100A9,92 which contributes to the occurrence and development of tumors. Compared with the corresponding TLR ligand, LPS from P. gingivalis showed weak M1-Mφ and M2-Mφ and induced the release of inflammatory cytokines, particularly TNF-α from M1-Mφ and IL-10 from M2-Mφ.93
The co-inhibitory receptor TIGIT (T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain) is expressed on both T cells and NK cells, and mediates tumor immunosuppression.94 F. nucleatum binds to TIGIT receptors via Fap2, which inhibits the activity of tumor infiltrating lymphocytes and NK cells.95 However, the combination of F. nucleatum and CEACAM1 is not FaP2-dependent.96 CEACAM1 is one of the carcinoembryonic antigens associated cell adhesion molecules (CEACAMs), which acts as a receptor for a variety of immune cell subsets and is expressed on a variety of tumors, including CRC.97 F. nucleatum has been shown to colonize the gut and combined with CEACAM1, inhibited the function of T cells and NK cells, and reduced the levels of IFN-γ and CD107a degranulation. Notably, the wild-type FN726 significantly inhibited IFN-γ and CD107a degranulation compared with the FaP2-deficient mutant FNK50.96 As FN726 is virulent through both TIGIT and CEACAM1, FNK50 can only be virulent through CEACAM1. This suggests that F. nucleatum inhibits anti-tumor immunity by activating the TIGIT receptor and CEACAM1, thus creating an immunosuppressive microenvironment for tumor cell avoidance (Figure 2).
Discussion
It has been widely recognized that intestinal and oral disorders play vital roles in a variety of diseases. To date, most studies on microbial related diseases have focused on individual organs, with less attention paid to microbial communication between differing organs. Accumulated evidence98,99 supports that oral microbes can transmitted to the intestinal tract and colonize, therefore altering the entire intestinal microbial ecosystem.
The oral cavity is an under-recognized and under-explored repository of the intestinal microbiome. Although traces of oral microbes appear in the feces of healthy individuals, oral pathogens do not settle in the gastrointestinal tract of healthy individuals. Two conditions are required for the colonization of oral pathogenic bacteria. One is microbial dysregulation due to the host gut disease, or CR provided by the gut microbiota is disrupted by antibiotics and the previously mentioned non-antibiotic drugs. The other is that oral pathogens, in some cases, exceed the threshold of digestive barrier resistance.27 Antibiotics not only eliminate pathogens, but also harm intestinal symbiotic bacteria.61 Based on this, more targeted antibiotics should be a direction of future efforts. Changes in the abundance of gut microbes should be closely watched during the administration of antibiotics, PPIs and antipsychotics, as this is a dangerous time for oral bacteria to spread and colonize within the gut. In addition, good oral hygiene and timely oral treatment can help prevent oral bacteria-mediated intestinal diseases, and even systemic diseases.
Crosstalk between the ectopic colonized oral pathogens and intestinal microbes have a particular affinity for tumorigenesis in gastrointestinal malignant diseases, including CRC, pancreatic,100 and liver cancers.101 It has been proposed that some disease models have helped to elucidate the dynamic changes of intestinal microbiome disorders in relation to the occurrence and progression of CRC, among which the driver-passenger model is the most classic.102 The “driver” bacteria have the potential to serve as CRC initiators, and eventually give way to “passenger” bacteria that facilitate or prevent tumorigenesis. Previous research has shown that F. nucleatum, P. micra has the potential to be a passenger in this process, while P. gingivalis has the potential to be a driver.22 This therefore may have the capacity to be an indicator of CRC. Several metagenomics sequencing results showed that intestinal microflora could be used as a potential marker for early CRC. Oral pathogens, such as F. nucleatum, P. micra,47 P. gingivalis,103 also had the ability to differentiate healthy people from those with CRC. With the development of sequencing technology, the use of oral bacteria as biomarkers for CRC detection will become possible. Studies have shown that fecal microbiota detection, combined with the fecal occult blood test, improves the diagnostic performance of CRC.104 Larger, more comprehensive cohort studies are needed in the future to validate the predictive value of oral bacteria for CRC.
A subset of CRC arises from chronic gut inflammation, for example, Crohn’s disease and ulcerative colitis.76 It is characterized by over-activation and recruitment of immune cells that produce inflammatory cytokines (TNF-α, IL-16, and IL-1β).105 Coincidentally, oral pathogens colonized in the intestinal tract activate the inflammasome and the NF-κB cascade pathway, inducing the release of pro-inflammatory cytokines, which creates an inflammatory microenvironment conducive to CRC. Immune evasion is a hallmark of cancer,106 and surprisingly, bacteria with oral to intestinal translocations are also involved in the construction of tumor immunosuppressive microenvironments by activating the host’s innate immune response. Perhaps future therapeutic strategies should focus on preventing the initiation and continuation of the inflammatory cascade before intestinal damage occurs. Attention should also be paid to immunotherapy of microbiological-correlated receptors. For example, TIGIT inhibitors have been used in combination with PD-1/PD-L1 inhibitors to achieve better tumor inhibition.107 Recently, prebiotics such as spores of Ganoderma Lucidum and probiotics such as Bifidobacterium breve have been found to reduce inflammation-mediated tumorigenesis and improve anti-tumor immunity, respectively.108 However, more clinical trials are needed in the future to evaluate the efficacy of probiotics and prebiotics in regulating tumor inflammatory microenvironment and improving tumor immunosuppressive microenvironment, so as to develop personalized probiotics and prebiotics treatment regimens and provide new therapeutic insights for CRC patients.
Conclusion
To sum up, oral bacteria colonize the gut and induce intestinal inflammation and immunosuppressive microenvironment, which are involved in the tumorigenesis and development of CRC. Comprehensive understanding of the contribution of the oral- intestinal microbiome axis in CRC provides benefits for early diagnosis and therapeutic strategies. Therefore, an in-depth study of microbial communication networks between organs will broaden the field of vision for the treatment and defense of microbial related diseases.
Acknowledgments
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn/) for the expert linguistic services provided.
Author Contributions
Sisi Mo and Haiming Ru share first authorship. All authors made significant contributions to conception and design, acquisition of data, or analysis and interpretation of data; participated in drafting the article or revising it critically; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for the contents of the article.
Funding
This work was supported by the National Natural Science Foundation of China (82160495 and 81973533), China Postdoctoral Science Foundation (No. 2019M653812XB), Guangxi University High-level Innovation Team and the Project of Outstanding Scholars Program (2019AC03004), and Guangxi Science and Technology Project (AD19245197).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lewis CM
2. Kitamoto S, Nagao-Kitamoto H, Hein R, Schmidt TM, Kamada N. The bacterial connection between the oral cavity and the gut diseases. J Dent Res. 2020;99(9):1021–1029. doi:10.1177/0022034520924633
3. Pandya G, Kirtonia A, Singh A, et al. A comprehensive review of the multifaceted role of the microbiota in human pancreatic carcinoma. Semin Cancer Biol. 2021. doi:10.1016/j.semcancer.2021.05.027
4. Sureda A, Daglia M, Argüelles Castilla S, et al. Oral microbiota and Alzheimer’s disease: Do all roads lead to Rome? Pharmacol Res. 2020;151(104582):104582. doi:10.1016/j.phrs.2019.104582
5. Hunt RH, Camilleri M, Crowe SE, et al. The stomach in health and disease. Gut. 2015;64(10):1650–1668. doi:10.1136/gutjnl-2014-307595
6. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15(2):111–128. doi:10.1038/nrgastro.2017.119
7. Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, Kuijper EJ. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev. 2019;83(3):e00007–19. doi:10.1128/mmbr.00007-19
8. Soderholm AT, Pedicord VA. Intestinal epithelial cells: at the interface of the microbiota and mucosal immunity. Immunology. 2019;158(4):267–280. doi:10.1111/imm.13117
9. Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279(1):90–105. doi:10.1111/imr.12563
10. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69(8):1510–1519. doi:10.1136/gutjnl-2019-320204
11. Brown EM, Kenny DJ, Xavier RJ. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol. 2019;37(1):599–624. doi:10.1146/annurev-immunol-042718-041841
12. Byrd KM, Gulati AS. The “Gum-Gut” axis in inflammatory bowel diseases: a hypothesis-driven review of associations and advances. Front Immunol. 2021;12:620124. doi:10.3389/fimmu.2021.620124
13. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN Estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
14. Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732. doi:10.1038/s41575-019-0189-8
15. Murphy N, Moreno V, Hughes DJ, et al. Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol Aspects Med. 2019;69:2–9. doi:10.1016/j.mam.2019.06.005
16. Yang L, Wang S, Lee JJ, et al. An enhanced genetic model of colorectal cancer progression history. Genome Biol. 2019;20(1):168. doi:10.1186/s13059-019-1782-4
17. Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585(7826):509–517. doi:10.1038/s41586-020-2729-3
18. Grazioso TP, Brandt M, Djouder N. Diet, microbiota, and colorectal cancer. iScience. 2019;21:168–187. doi:10.1016/j.isci.2019.10.011
19. Imai S, Ooki T, Murata-Kamiya N, et al. Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe. 2021;29(6):941–958.e10. doi:10.1016/j.chom.2021.04.006
20. Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454–1463. doi:10.1136/gutjnl-2017-314814
21. Hong J, Guo F, Lu SY, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2020;70(11):2123–2137. doi:10.1136/gutjnl-2020-322780
22. Gao R, Zhu Y, Kong C, et al. Alterations, interactions, and diagnostic potential of gut bacteria and viruses in colorectal cancer. Front Cell Infect Microbiol. 2021;11:657867. doi:10.3389/fcimb.2021.657867
23. Blasco-Baque V, Garidou L, Pomié C, et al. Periodontitis induced by Porphyromonas gingivalis drives periodontal microbiota dysbiosis and insulin resistance via an impaired adaptive immune response. Gut. 2017;66(5):872–885. doi:10.1136/gutjnl-2015-309897
24. Esberg A, Haworth S, Kuja-Halkola R, Magnusson PKE, Johansson I. Heritability of oral microbiota and immune responses to oral bacteria. Microorganisms. 2020;8(8):1126. doi:10.3390/microorganisms8081126
25. Kim YK, Shin C. The microbiota-gut-brain axis in neuropsychiatric disorders: pathophysiological mechanisms and novel treatments. Curr Neuropharmacol. 2018;16(5):559–573. doi:10.2174/1570159x15666170915141036
26. Tranah TH, Edwards LA, Schnabl B, Shawcross DL. Targeting the gut-liver-immune axis to treat cirrhosis. Gut. 2021;70(5):982–994. doi:10.1136/gutjnl-2020-320786
27. Kitamoto S, Nagao-Kitamoto H, Jiao Y, et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182(2):447–462. e14. doi:10.1016/j.cell.2020.05.048
28. Mark Welch JL, Ramírez -puebla ST, Borisy GG. Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe. 2020;28(2):160–168. doi:10.1016/j.chom.2020.07.009
29. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi:10.1038/nature11234.
30. Utter DR, Borisy GG, Eren AM, Cavanaugh CM, Mark Welch JL. Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. Genome Biol. 2020;21(1):293. doi:10.1186/s13059-020-02200-2
31. Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9(5):488–500. doi:10.1007/s13238-018-0548-1
32. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16(12):745–759. doi:10.1038/s41579-018-0089-x
33. Dinakaran V, Mandape SN, Shuba K, et al. Identification of specific oral and gut pathogens in full thickness colon of colitis patients: implications for colon motility. Front Microbiol. 2018;9:3220. doi:10.3389/fmicb.2018.03220
34. Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76(3):473–493. doi:10.1007/s00018-018-2943-4
35. Magne F, Gotteland M, Gauthier L, et al. The Firmicutes/Bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12(5):1474. doi:10.3390/nu12051474
36. Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the Firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 2020;8(11):1715. doi:10.3390/microorganisms8111715
37. Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi:10.1038/nature09944
38. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20(2):145–155. doi:10.1038/nn.4476
39. Gomes AC, Hoffmann C, Mota JF. The human gut microbiota: metabolism and perspective in obesity. Gut Microbes. 2018;9(4):308–325. doi:10.1080/19490976.2018.1465157
40. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705–715. doi:10.1016/j.chom.2018.05.012
41. Salazar N, Valdés-Varela L, González S, Gueimonde M, de Los Reyes- Gavilán CG. Nutrition and the gut microbiome in the elderly. Gut Microbes. 2017;8(2):82–97. doi:10.1080/19490976.2016.1256525
42. Pickard JM, Zeng MY, Caruso R, Nunez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279(1):70–89. doi:10.1111/imr.12567
43. Oliveira RA, Ng KM, Correia MB, et al. Klebsiella michiganensis transmission enhances resistance to Enterobacteriaceae gut invasion by nutrition competition. Nat Microbiol. 2020;5(4):630–641. doi:10.1038/s41564-019-0658-4
44. Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi:10.1038/ni.2608
45. Pickard JM, Nunez G. Pathogen colonization resistance in the gut and its manipulation for improved health. Am J Pathol. 2019;189(7):1300–1310. doi:10.1016/j.ajpath.2019.03.003
46. Yang Y, Misra BB, Liang L, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9(14):4101–4114. doi:10.7150/thno.35186
47. Dai Z, Coker OO, Nakatsu G, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6(1):70. doi:10.1186/s40168-018-0451-2
48. Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol Med. 2017;23(1):18–30. doi:10.1016/j.molmed.2016.11.004
49. Russo E, Bacci G, Chiellini C, et al. Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: a pilot study. Front Microbiol. 2017;8:2699. doi:10.3389/fmicb.2017.02699
50. Komiya Y, Shimomura Y, Higurashi T, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68(7):1335–1337. doi:10.1136/gutjnl-2018-316661
51. Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi:10.1038/s41591-019-0458-7
52. Kaci G, Goudercourt D, Dennin V, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol. 2014;80(3):928–934. doi:10.1128/aem.03133-13
53. Said HS, Suda W, Nakagome S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014;21(1):15–25. doi:10.1093/dnares/dst037
54. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi:10.1016/j.chom.2013.07.007
55. Yang J, Li D, Yang Z, et al. Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut Microbes. 2020;11(4):918–929. doi:10.1080/19490976.2020.1712986
56. Allali I, Delgado S, Marron PI, et al. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes. 2015;6(3):161–172. doi:10.1080/19490976.2015.1039223
57. Koliarakis I, Messaritakis I, Nikolouzakis TK, Hamilos G, Souglakos J, Tsiaoussis J. Oral bacteria and intestinal dysbiosis in colorectal cancer. Int J Mol Sci. 2019;20(17):4146. doi:10.3390/ijms20174146
58. Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J Oral Microbiol. 2019;11(1):1586422. doi:10.1080/20002297.2019.1586422
59. Casasanta MA, Yoo CC, Udayasuryan B, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. 2020;13(641):eaba9157. doi:10.1126/scisignal.aba9157
60. Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi:10.1126/science.aal5240
61. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65(11):1906–1915. doi:10.1136/gutjnl-2016-312297
62. Scott NA, Andrusaite A, Andersen P, et al. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med. 2018;10(464):eaao4755. doi:10.1126/scitranslmed.aao4755
63. Atarashi K, Suda W, Luo C, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359–365. doi:10.1126/science.aan4526
64. Jackson MA, Goodrich JK, Maxan ME, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65(5):749–756. doi:10.1136/gutjnl-2015-310861
65. Takashima S, Tanaka F, Kawaguchi Y, et al. Proton pump inhibitors enhance intestinal permeability via dysbiosis of gut microbiota under stressed conditions in mice. Neurogastroenterol Motil. 2020;32(7):e13841. doi:10.1111/nmo.13841
66. Morgan AP, Crowley JJ, Nonneman RJ, et al. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One. 2014;9(12):e115225. doi:10.1371/journal.pone.0115225
67. Cussotto S, Strain CR, Fouhy F, et al. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology. 2019;236(5):1671–1685. doi:10.1007/s00213-018-5006-5
68. Sokol H, Seksik P, Rigottier-Gois L, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12(2):106–111. doi:10.1097/01.MIB.0000200323.38139.c6
69. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi:10.1186/gb-2012-13-9-r79
70. Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16(1):38–51. doi:10.1080/15548627.2019.1635384
71. Kitamoto S, Alteri CJ, Rodrigues M, et al. Dietary L-serine confers a competitive fitness advantage to Enterobacteriaceae in the inflamed gut. Nat Microbiol. 2020;5(1):116–125. doi:10.1038/s41564-019-0591-6
72. Stecher B, Conway T, Cohen P. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr. 2015;3(3). doi:10.1128/microbiolspec.MBP-0008-2014
73. Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67(6):1024–1032. doi:10.1136/gutjnl-2017-314281
74. Gourley CR, Negretti NM, Konkel ME. The food-borne pathogen Campylobacter jejuni depends on the AddAB DNA repair system to defend against bile in the intestinal environment. Sci Rep. 2017;7(1):14777. doi:10.1038/s41598-017-14646-9
75. Boll EJ, Nielsen LN, Krogfelt KA, Struve C. Novel screening assay for in vivo selection of Klebsiella pneumoniae genes promoting gastrointestinal colonisation. BMC Microbiol. 2012;12(1):201. doi:10.1186/1471-2180-12-201
76. Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140(6):1807–1816. doi:10.1053/j.gastro.2011.01.057
77. Wang G, Huang S, Wang Y, et al. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci. 2019;76(20):3917–3937. doi:10.1007/s00018-019-03190-6
78. Bauer C, Duewell P, Mayer C, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59(9):1192–1199. doi:10.1136/gut.2009.197822
79. Peng C, Ouyang Y, Lu N, Li LN. The NF-κB signaling pathway, the microbiota, and gastrointestinal tumorigenesis: recent advances. Front Immunol. 2020;11:1387. doi:10.3389/fimmu.2020.01387
80. Engevik MA, Danhof HA, Ruan W, et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio. 2021;12(2):e02706–02720. doi:10.1128/mBio.02706-20
81. Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21(2):223–244. doi:10.1038/cr.2011.13
82. Jia YP, Wang K, Zhang ZJ, et al. TLR2/TLR4 activation induces Tregs and suppresses intestinal inflammation caused by Fusobacterium nucleatum in vivo. PLoS One. 2017;12(10):e0186179. doi:10.1371/journal.pone.0186179
83. Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-н╨B, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–866.e24. doi:10.1053/j.gastro.2016.11.018
84. Shi C, Yang Y, Xia Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65(9):1470–1481. doi:10.1136/gutjnl-2014-308455
85. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi:10.1038/nrc3611
86. Atanasova K, Lee J, Roberts J, Lee K, Ojcius DM, Yilmaz Ö. Nucleoside-diphosphate-kinase of p. gingivalis is secreted from epithelial cells in the absence of a leader sequence through a pannexin-1 interactome. Sci Rep. 2016;6(1):37643. doi:10.1038/srep37643
87. Wang X, Jia Y, Wen L, et al. Porphyromonas gingivalis promotes colorectal carcinoma by activating the hematopoietic NLRP3 inflammasome. Cancer Res. 2021;81(10):2745–2759. doi:10.1158/0008-5472.CAN-20-3827
88. Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect Immun. 2014;82(1):112–123. doi:10.1128/iai.00862-13
89. Khan AA, Khan Z, Warnakulasuriya S. Cancer-associated toll-like receptor modulation and insinuation in infection susceptibility: association or coincidence? Ann Oncol. 2016;27(6):984–997. doi:10.1093/annonc/mdw053
90. Burgueño JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17(5):263–278. doi:10.1038/s41575-019-0261-4
91. Chen T, Li Q, Wu J, et al. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol Immunother. 2018;67(10):1635–1646. doi:10.1007/s00262-018-2233-x
92. Hu L, Liu Y, Kong X, et al. Fusobacterium nucleatum facilitates M2 macrophage polarization and colorectal carcinoma progression by activating TLR4/NF-κB/S100A9 cascade. Front Immunol. 2021;12:658681. doi:10.3389/fimmu.2021.658681
93. Holden JA, Attard TJ, Laughton KM, Mansell A, O’Brien-Simpson NM, Reynolds EC. Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect Immun. 2014;82(10):4190–4203. doi:10.1128/iai.02325-14
94. Zhang Q, Bi J, Zheng X, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol. 2018;19(7):723–732. doi:10.1038/s41590-018-0132-0
95. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi:10.1016/j.immuni.2015.01.010
96. Gur C, Maalouf N, Shhadeh A, et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology. 2019;8(6):e1581531. doi:10.1080/2162402x.2019.1581531
97. Kim WM, Huang YH, Gandhi A, Blumberg RS. CEACAM1 structure and function in immunity and its therapeutic implications. Semin Immunol. 2019;42:101296. doi:10.1016/j.smim.2019.101296
98. Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6(1):8727. doi:10.1038/ncomms9727
99. Uchino Y, Goto Y, Konishi Y, et al. Colorectal cancer patients have four specific bacterial species in oral and gut microbiota in common-a metagenomic comparison with healthy subjects. Cancers. 2021;13(13):3332. doi:10.3390/cancers13133332
100. Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–127. doi:10.1136/gutjnl-2016-312580
101. Li D, Xi W, Zhang Z, et al. Oral microbial community analysis of the patients in the progression of liver cancer. Microb Pathog. 2020;149:104479. doi:10.1016/j.micpath.2020.104479
102. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–582. doi:10.1038/nrmicro2819
103. Guilloux CA, Lamoureux C, Beauruelle C, Héry-Arnaud G. Porphyromonas: a neglected potential key genus in human microbiomes. Anaerobe. 2021;68:102230. doi:10.1016/j.anaerobe.2020.102230
104. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. doi:10.15252/msb.20145645
105. Singh UP, Singh NP, Murphy EA, et al. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine. 2016;77:44–49. doi:10.1016/j.cyto.2015.10.008
106. Sasidharan Nair V, Toor SM, Taha RZ, Shaath H, Elkord E. DNA methylation and repressive histones in the promoters of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, PD-L1, and galectin-9 genes in human colorectal cancer. Clin Epigenetics. 2018;10(1):104. doi:10.1186/s13148-018-0539-3
107. Johnston RJ, Comps-Agrar L, Hackney J, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26(6):923–937. doi:10.1016/j.ccell.2014.10.018
108. Li Y, Liu H, Qi H, et al. Probiotic fermentation of Ganoderma lucidum fruiting body extracts promoted its immunostimulatory activity in mice with dexamethasone-induced immunosuppression. Biomed Pharmacother. 2021;141:111909. doi:10.1016/j.biopha.2021.111909
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.