Back to Journals » International Journal of Nanomedicine » Volume 12
Optimization and physicochemical characterization of a cationic lipid-phosphatidylcholine mixed emulsion formulated as a highly efficient vehicle that facilitates adenoviral gene transfer
Authors Kim SY, Lee SJ, Kim JK, Choi HG, Lim SJ
Received 19 July 2017
Accepted for publication 14 September 2017
Published 9 October 2017 Volume 2017:12 Pages 7323—7335
DOI https://doi.org/10.2147/IJN.S146785
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Soo-Yeon Kim,1,2 Sang-Jin Lee,2 Jin-Ki Kim,3 Han-Gon Choi,3 Soo-Jeong Lim1
1Department of Bioscience and Bioengineering, Sejong University, Seoul, Kwangjin-gu, Seoul, 2Immunotherapeutics Branch, Research Institute, National Cancer Center, Ilsandong-gu, Goyang-si, Gyeonggi-do, 3College of Pharmacy & Institute of Pharmaceutical Science and Technology, Hanyang University, Sangnok-gu, Ansan, Republic of Korea
Abstract: Cationic lipid-based nanoparticles enhance viral gene transfer by forming electrostatic complexes with adenoviral vectors. We recently demonstrated the superior complexation capabilities of 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) emulsion compared with a liposomal counterpart but the cytotoxicity of DOTAP emulsions remained a challenge. The present study is aimed at formulating an emulsion capable of acting as a highly effective viral gene transfer vehicle with reduced cytotoxicity and to physicochemically characterize the structures of virus-emulsion complexes in comparison with virus–liposome complexes when the only difference between emulsions and liposomes was the presence or absence of inner oil core. The emulsion formulation was performed by 1) reducing the content of DOTAP while increasing the content of zwitterionic lipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and 2) optimizing the oil content. The complexation capability of formulated DOTAP:DMPC mixed emulsions was similar to those of emulsions containing DOTAP alone while displaying significantly lower cytotoxicity. The complexation capabilities of the DOTAP:DMPC mixed emulsion were serum-compatible and were monitored in a variety of cell types, whereas its liposomal counterpart was totally ineffective. Characterization by scanning electron microscopy, transmission electron microscopy, atomic force microscopy, and dynamic light scattering studies indicated that the optimized emulsions spontaneously surrounded the virus particles to generate emulsions that encapsulated the viral particles, whereas viral particles merely attached to the surfaces of the counterpart liposomes to form multiviral aggregates. Overall, these studies demonstrated that optimized DOTAP:DMPC mixed emulsions are potentially useful for adenoviral gene delivery due to less cytotoxicity and the unique ability to encapsulate the viral particle, highlighting the importance of nanoparticle formulation.
Keywords: cationic lipid, oil, emulsion, adenovirus, gene delivery
Introduction
Adenovirus (Ad) is an 80–100 nm non-enveloped virus comprising a 36 kb linear double-stranded DNA genome and core proteins surrounded by capsid proteins. The use of adenovirus-derived vectors in gene delivery has several clinically useful features, including a high gene transfer efficiency in both dividing and non-dividing cells and easy production of high-titer Ad stocks;1 thus, these vectors are currently one of the most widely used means for delivering genes.2
Ad entry into cells is initiated by the virus attachment, via the adenoviral fiber knob, to a primary adhesion receptor, the coxsackievirus and Ad receptor (CAR), present on the target cell membrane. The Ad rapidly escapes the endosome and enters the nucleus under the direction of inherent Ad proteins after internalization by endocytosis;3 therefore, the gene transfer efficiencies of adenovirus-derived vectors are often determined by the CAR expression level on the surface of a target cell. Unfortunately, CAR receptors are not significantly expressed by certain types of cells, including primary cancer cells, differentiated endothelial and epithelial cells, and mesenchymal or hematopoietic stem cells,4–6 which hampers efficient gene transfer in these cells.
Numerous efforts have attempted to overcome the limitations of adenoviral vector delivery posed by low CAR expression levels in many cell types. Several studies have demonstrated that this limitation may be overcome by electrostatically complexing adenoviruses with cationic vehicles, particularly cationic lipid-based nanoparticles, that exploit the anionic properties of the outer surfaces (eg, hexons) of the adenoviral particles. Simple mixing of adenoviral particles and cationic nanoparticles spontaneously produces complexes, and the Ad–nanoparticle complexes facilitate viral attachment to negatively charged cell membranes and increase gene transfer in both CAR-positive and CAR-negative cells.7–10
The composition of a nanoparticle determines the ability of that nanoparticle to act as a carrier of chemicals and biologics.11 The type and content of a cationic lipid and other components used to form cationic lipid-based nanoparticles can affect the surface charge, size, and structure of the nanoparticles, thereby varying the strength of the interactions between nanoparticles and viral particles. The strength of interaction within a complex is a major determinant of the efficacy of complex-mediated viral gene transfer, particularly under complex-destabilizing conditions, such as those present in serum.12,13 A very limited number of studies, however, have addressed this issue in adenoviral gene delivery. Compared with liposomes containing 1,2-dioleyloxy-3-trimethylammonium propane chloride as a cationic lipid (Lipofectin®), dioleoyltrimethylaminopropane (DOTAP)-containing liposomes were found to be more effective in enhancing adenovirus-mediated transgene expression,7 emphasizing the importance of the of cationic lipid’s chemical structure.14 DOTAP liposomes supplemented with PEGylated phospholipid turned out to be more effective compared with their non-PEGylated counterpart liposomes, probably due to the increased stability of the adenovirus-PEGylated liposome complexes.7 Liposomes composed of DOTAP and cholesterol (CHOL) formed more tightly associated adenoviral complexes compared with those composed of DOTAP and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), resulting in a greater complexing effect, that was thought to arise from the strong affinity between the Ad and CHOL.15,16 Taken together, these findings highlight the importance of the composition of a cationic nanoparticle in determining its complexation effects on viral gene transfer.
Our recent work demonstrated that DOTAP emulsions comprising a Lipidol® (iodized oil) inner oil core stabilized by a DOTAP:CHOL:1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-5000] (PEG-PE) mixture were more capable of enhancing adenovirus-mediated gene transfer compared with liposomal counterparts. The only difference between these 2 formulations was the presence or absence of the inner oil core.16 In this work, emulsion complexation was found to shield the negative surface charges on the adenoviral particles more effectively than liposomes, and this shielding increased the gene transfer capabilities of the emulsions. Prior studies by others have shown that the structures of virus–nanoparticle complexes may differ substantially, depending on the composition of the nanoparticle: mixing an Ad with DOTAP:CHOL liposomes resulted in virus envelopment within the lipid layers,17 whereas mixing with 2,3-dioleoyloxy-N-[2-(spermine-carboxamido)ethyl]-N,N-dimethyl-1-propanaminium:DOPE liposomes (Lipofectamine®) formed virus–liposome aggregates without any evidence of virus encapsulation.18 In this background, the present study was undertaken to physicochemically characterize the structures of virus–emulsion complexes in comparison with those of virus–liposome complexes to explore why emulsions are more effective than liposomes.
Cationic lipid-based nanoparticles, including DOTAP-based ones, are generally cytotoxic,19 although the cytotoxicity tends to be lower than that of cationic polymer-based nanoparticles. The nanoparticle cytotoxicity arises from the cationic lipids,20,21 which generate intracellular reactive oxygen species that lead to a calcium influx into cells, which destabilizes the cellular plasma membrane upon transfer from the nanoparticles into the membrane and/or inhibits Na+/K+ pump activity in the cells by interacting with the cation-binding site of the pump.22,23 Efforts to formulate nanoparticles with lower cationic lipid contents while preserving the binding properties to viruses can be an effective strategy to balance the cytotoxicity and complexation efficacy. Others have reported that zwitterionic lipids, particularly phosphatidylcholines, interact with and specifically bind to hexons, the major capsid proteins of adenoviruses.24,25 With this in mind, we sought to optimize the cationic lipid:phosphatidylcholine mixed nanoparticle formula as the second objective of the present study.
Materials and methods
Reagents and materials
DOTAP, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and PEG-PE were purchased from Avanti Polar Lipids (Alabaster, AL, USA). CHOL was purchased from Sigma-Aldrich (St Louis, MO, USA). Lipiodol®, an iodinated ethyl ester of poppy seed oil, was purchased from Guerbet Antre (Aulnay-sous-Bois, France).
The recombinant adenoviral Ad encoding green fluorescence protein (Ad-GFP) (replication-defective Ad type 5) viruses under the control of cytomegalovirus promoter were constructed in E1/E3-deleted RightZap1.2 vector (OD260, Boise, ID, USA) and propagated in a permissive 293 cell line. Produced Ad particles were purified by ultra-centrifugation through cesium chloride gradients. Viral titers were determined by absorbance at 260 nm. Plaque-forming unit (pfu) was calculated by determining the maximal dilution factor to be able to lyze 293 cells in a 96-well plate. The viral stock was kept frozen at −80°C until use.
All other chemicals were of reagent grade and used without further purification.
Cell line and culture
Mouse B16-F10 melanoma cells, human H460 non-small cell lung cancer cells and HCT116 colon cancer cells were obtained from the Korean Cell Line Bank (Seoul, Korea). EA.hy926 human umbilical vein endothelial cells were obtained from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in DMEM (B16-F10, EA.hy926) or RPMI 1640 (H460, HCT116) medium (Welgene, Daegu, Korea) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY, USA) and 100 units/mL of each of penicillin and streptomycin. Cells were grown in incubators in a humid atmosphere of 95% air and 5% CO2 at 37°C.
Preparation of liposomes and emulsions
Liposomes and emulsions were prepared as described in our earlier studies.10,16 Briefly, a total 7 μmol of lipid mixture composed of DOTAP, DMPC, CHOL and PEG-PE was dissolved together in 2 mL of tert-butyl alcohol. After rapid freezing at −80°C, mixtures were subjected to freeze-drying by freeze dryer (FDU-1200, Eyela, Tokyo, Japan). Lipid cakes obtained after lyophilization were hydrated with 5% dextrose dissolved in distilled water of 1 mL. For preparing emulsions, Lipiodol was added to the lipid cakes prior to hydration. The hydrated lipid dispersion was briefly vortexed and sonicated in bath-type sonicator for 2 h at 37°C. The prepared formulations were stored at 4°C until use.
Characterization by dynamic light scattering
Mean particle sizes of liposomes, emulsions or their complexes with Ad were determined using a fiber-optic particle analyzer (FPAR-1000, Otsuka Electronics, Osaka, Japan). Prior to measurement, samples were diluted with filtered 5% dextrose solution. In the diluted sample, 28 nmol of total lipids complexed with 8×107 pfu were present in 1 mL of 5% dextrose solution. The system was used in the auto-measuring mode. Particle size analysis data were evaluated using CONTIN program provided by the manufacturer.
Zeta potential was determined using a Zen 600 zetasizer (Malvern, England). Prior to measurement, samples were diluted to 1 mL with deionized water. Default instrument settings and automatic analysis were used for all measurements. Each measurement was carried out in triplicate. In the case of samples containing Ad-GFP, Ad-GFP at a concentration of 2×109 pfu was included in the samples.
Adenoviral gene transfer
Viral gene transfer was performed as described previously.16 Briefly, cells were seeded in 12-well tissue culture plates at a density of 60,000 cells/well, kept in a CO2 incubator at 37°C and used when 70%–80% confluent. Complexes of Ad-GFP and emulsions (at the ratio of 3.5 nmol total lipid/107 pfu Ad-GFP) were made by mixing adenoviral vectors and emulsions using gentle pipette tip aspiration, followed by incubation for 20 min at room temperature. Cells were washed with PBS, and virus-emulsion (or liposome) complexes were added together with 400 L medium with or without serum supplementation. After 4h incubation in a CO2 incubator at 37°C, cells were washed with PBS to remove the complexes, and 1 mL fresh culture media supplemented with 10% serum was added. Cells were then incubated for an additional 36h before assessing GFP expression.
Evaluation of transgene expression
After incubation, cells were harvested by using a scraper. Harvested cells were washed twice with PBS and suspended in fresh medium supplemented with 10% serum. Cells were fixed with 1 mL 1% paraformaldehyde for 30 min at 4°C. After brief centrifugation, the supernatant was discarded and fixed cells were resuspended with PBS. The expression of GFP in suspended cells was determined by using Beckman Coulter flow cytometer (Beckman Coulter Korea, Seoul, Korea) using Cell Quest program (BD Bioscience, San Jose, CA, USA). Ten thousand fluorescent events per sample were acquired using a 530/15 band pass filter for the GFP signal obtained with fluorescence emission centered at 530 nm.
To evaluate the transgene expression by confocal microscopy, cells were seeded on a cover glass placed in 12-well plates and then infected with Ad complexes by using the same protocol, as aforedescribed. After removing the culture media, cells were washed 3 times with cold PBS and fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were then washed 3 times with cold PBS. Mounting solution (Dako Korea, Seoul, Korea) was dropped on a slide glass and the cover glass was put on the slide glass to contact the mounting solution. The fixed cells were observed by Leica TCS SP5 Confocal Laser Scanning Microscope (Wetzlar, Germany). Fluorescence imaging of each sample was obtained using 488 nm excitation line and a band-pass BP495-555 emission filter.
Characterization by electron microscopy
The morphology of Ad–emulsion or Ad–liposome complexes was examined by negative stain transmission electron microscopy (TEM) and field-emission scanning electron microscopy (FESEM).
Prior to FESEM (AURIGA® Carl Zeiss, Oberkochen, Germany) analysis, 20 μL of samples containing complexes was deposited on the surface of freshly cleaved mica, and samples were allowed to adsorb for 15 min. Unbound samples were removed by washing with distilled water. Samples were then dried under a nitrogen stream. Samples were coated with a thin layer of gold (10 nm) before FESEM observation. Images were taken at accelerating voltage of 2 kV.
To examine the morphology of complexes by TEM, 5 μL of each sample was applied to carbon-coated grids that had been glow-discharged for 3 min in air and immediately (~5 sec) negatively stained using 1% uranyl acetate. Excess stain was removed, samples were allowed to air-dry completely, and micrographs were taken using a Tecnai G2 Spirit (FEI Company, Hilsbro, OR, USA).
Atomic force microscopy (AFM)
A 20 mL suspension containing Ad–liposome or Ad–emulsion complexes was deposited on the surface of freshly cleaved mica, and samples were allowed to adsorb for 15 min. Unbound samples were removed by washing with distilled water. Samples were then dried under a nitrogen stream. Imaging was carried out in Tapping Mode using a Multimode AFM (Park Systems, Suwon, Korea), Z scanner, XE-100 controller, XEI control software and a silicon tapping tip (PPP-NCHR, with aluminum coating detector) of 30 nm curvature radius, mounted on a tapping-mode silicon cantilever with a typical resonant frequency of 371 kHz and a force constant of 42 N/m, to image 5×5 μm square areas of the mica surface with a resolution of 256×256 pixels. All AFM imaging was performed in air.
MTT assay
B16-F10 melanoma cells seeded in 96-well or 12-well plates were treated with serial dilutions of varying emulsion formulations with or without Ad complexation. After incubation in a CO2 incubator at 37°C, cell viability was measured using MTT assay. Media were replaced by 100 μL fresh media containing 0.5 mg/mL of MTT (Amresco, Solon, OH, USA). After 4h incubation in a CO2 incubator at 37°C, MTT-containing media were removed and generated formazan crystals in cells were solubilized by adding 100 μL of dimethyl sulfoxide to each well. The absorbance was determined using an EL 800 Universal Microplate Reader (BioTek, Winooski, VT, USA) at 540 nm.
Statistical analysis
Statistically significant differences between values obtained in different compositions of emulsions (liposomes) or under different experimental conditions were determined using 2-tailed unpaired Student’s t-tests.
Results and discussion
Effect of DOTAP content on adenoviral gene transfer efficiency
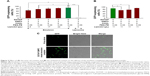
In a previous study, cationic emulsions used to form adenoviral particle complexes were prepared with a lipid mixture containing a 6:6:0.06 mixture of DOTAP:CHOL:PEG-PE. Ad complex of emulsion prepared with this mixture and Ads displayed a very high transgene expression efficiency.16 In an effort to prepare a highly effective vehicle with a low cytotoxicity by substituting some DOTAP with DMPC, while the ratio of DOTAP plus DMPC:CHOL:PEG-PE was fixed at 6:6:0.01, were prepared. The properties of these emulsion complexes were then evaluated by measuring the percentage of GFP-expressing cells after gene transfer with a fixed dose of an Ad-GFP pre-complexed with the various emulsions. Figure 1A indicates that adenoviral complexation with emulsions formed with 4:2 or 6:0 DOTAP:DMPC mixtures produced comparable GFP expression results, increasing GFP expression by factors of 16.9 and 17.7 compared with the virus alone. Emulsions prepared with a 3:3 mixture of DOTAP:DMPC increased the number of GFP-expressing cells to a slightly lower degree (by a factor of 15.2 compared with the virus alone), and emulsions prepared with a 2:4 mixture of DOTAP:DMPC were totally ineffective: the GFP expression was barely detectable, similar to the results obtained from the Ad-GFP alone (<5% in both cases). These data suggested that the emulsions composed of DOTAP plus DMPC, CHOL and PEG-PE in a ratio of 6:6:0.01 could form tight complexes with the adenoviral particles if ≥50% DOTAP were used in the DOTAP:DMPC mixture.
Substitution of the cationic lipid (DOTAP) with a zwitterionic lipid (DMPC) in the emulsions was expected to decrease the cationic surface charges in the emulsions. The relationship between the gene transfer enhancement effect and the surface charge on the emulsions was investigated by measuring the zeta potential of each emulsion as a function of the DOTAP:DMPC ratio. There was no tendency for the zeta potential values to change linearly with decreasing DOTAP content (between +57 and +69 mV) in the emulsions containing varying DOTAP content. Only the control DMPC:CHOL:PEG-PE emulsions exhibited highly negative surface charges due to the presence of PEG-PE and the absence of DOTAP (Figure 1B). These results indicated that the DOTAP surface densities in all DOTAP:DMPC mixed emulsions were comparable, despite the varying DOTAP contents, suggesting that only a fraction of the DOTAP molecules were exposed on the surfaces of the iodized oil emulsions. Some residual DOTAP molecules may have been buried inside the emulsions through partial dissolution in oil due to the hydrophobic interactions between the oil and the 2 unsaturated hydrocarbon chains of the DOTAP molecules. Increasing the DOTAP content may have increased the DOTAP content inside the emulsions. Despite having cationic surface charges similar to those of other DOTAP emulsions, the ineffective gene transfer results obtained from emulsions containing 2:4 DOTAP:DMPC suggested that the buried DOTAP molecules are also involved in achieving tight complexation with the adenoviral particles.
Effect of the oil and CHOL content of the emulsions on the adenoviral gene transfer efficiency
An earlier study showed that a higher Lipiodol content in DOTAP emulsions enhanced adenoviral transgene expression up to 11 μL/mL of Lipiodol.16 In an effort to reduce the cytotoxicity while preserving the effectiveness of the gene transfer vehicle, the Lipiodol content of 3:3 DOTAP:DMPC mixed emulsions was varied from 0 to 22 μL/mL (Figure 2A). Increasing the Lipiodol content enhanced the transgene expression in a linear fashion, and a 3:3 DOTAP:DMPC mixed emulsion with ≥17 μL Lipiodol produced GFP expression levels in cells comparable with those obtained from 6:0 DOTAP:DMPC emulsions prepared with 11 μL Lipiodol (p>0.05). By contrast, 3:3 DOTAP:DMPC mixed liposome complexes were totally ineffective in enhancing adenoviral gene transfer. These data indicated that the inner oil core played a critical role in achieving tight complexation between Ad and DOTAP:DMPC mixed nanoparticles.
CHOL is an important stabilizing component of cationic lipid-based nanoparticles; therefore, we sought to optimize the CHOL content in DOTAP:DMPC mixed emulsions. Fixing the DOTAP and Lipiodol contents at 3 and 11 μL/mL emulsion, respectively, we found that decreasing the DMPC content while increasing the CHOL content increased GFP expression in a CHOL content-dependent manner (Figure 2B). The percentage of GFP-expressing cells obtained using a 3:0:9:0.06 DOTAP:DMPC:CHOL:PEG-PE emulsion complexation was similar to that obtained using a 6:0:6:0.06 DOTAP:DMPC:CHOL:PEG-PE emulsion complexation (p>0.05). We speculate that the enhancement effect obtained upon incorporating CHOL into the emulsion may have arisen from the binding of CHOL to the adenoviral particles.15 Confocal laser microcopy confirmed the substantially higher GFP expression in B16-F10 cells infected with Ad-GFP complexed with a 3:3:6:0.06 DOTAP:DMPC:CHOL:PEG-PE emulsion containing 17 μL/mL Lipiodol, in contrast to the minimal expression obtained from Ad-GFP alone or from complexes with 3:3:6:0.06 DOTAP:DMPC:CHOL:PEG-PE liposomes (Figure 2C, data not shown for the liposomes). Taken together, 3:3:6:0.06 or 3:0:9:0.06 DOTAP:DMPC:CHOL:PEG-PE emulsions, prepared with a low cationic lipid content, effectively increased the adenoviral transgene expression, comparable with 6:0:6:0.06 DOTAP:DMPC:CHOL:PEG-PE emulsions. Based on literature reports, it is probable that quite strong interactions exist between the Ad and DMPC15 or between the Ad and CHOL,24,25 thereby complementing the electrostatic binding between DOTAP and adenoviral particles. Combined together with DOTAP:DMPC ratio data shown in Figure 1, it seems likely that even if a portion of DOTAP in emulsions is substituted with DMPC or CHOL, if >25% of DOTAP is included in the total lipid mixture (DOTAP:DMPC:CHOL:PEG-PE), emulsions are capable of enhancing the adenoviral gene transfer by complexation.
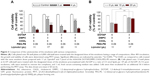
Evaluation of the cytotoxicity of the DOTAP:DMPC mixed emulsions
The cytotoxicities of 4 DOTAP or DOTAP:DMPC mixed emulsion formulations with comparable GFP expression enhancement effects (±80% GFP-expressing cells by complexation) were compared. After 48 h continuous incubation of B16-F10 cells with the emulsions, all 4 formulations inhibited cellular proliferation in an emulsion dose-dependent manner, but the extent of cytotoxicity differed among the formulations (Figure 3A). Compared with the DOTAP emulsions prepared with double the DOTAP content (emulsions with a 6:6:0.06 mixture of DOTAP:CHOL:PEG-PE and 11 μL/mL Lipiodol), the cytotoxicities of the DOTAP:DMPC mixed emulsions prepared using a 3:3:6:0.06 mixture of DOTAP:DMPC:CHOL:PEG-PE and 22 μL/mL Lipiodol were similar. DOTAP:DMPC mixed emulsions prepared with a 3:3:6:0.06 mixture of DOTAP:DMPC:CHOL:PEG-PE and 17 μL/mL Lipiodol were less cytotoxic at both the 2 and 4 μL emulsion doses (p<0.001), and those prepared with a 3:0:9:0.06 mixture of DOTAP:DMPC:CHOL:PEG-PE and 11 μL/mL Lipiodol were less cytotoxic only at a 2 μL dose (p<0.001).
The cationic lipid content in the nanoparticles is known to be a major factor determining the cytotoxicity of the cationic lipid-based nanoparticles.26 Our data suggest that the cytotoxicities of the cationic lipid–phosphatidylcholine mixed emulsions are affected by the inner oil and CHOL contents in addition to the cationic lipid content. With our current data, the mechanism causing the difference in cytotoxicity is unclear but the different composition of emulsions may affect the strength of the interactions between DOTAP and the cellular membranes, which may, in turn, destabilize the cellular plasma membrane and/or inhibit the activity of Na+/K+ pumps.22,23
The cytotoxicities of the DOTAP:DMPC mixed emulsions were compared with the cytotoxicity of a DOTAP-alone emulsion using the adenoviral gene transfer protocol. The growth and viability of cells were determined after cell transduction with Ad-GFP complexed with the DOTAP:DMPC mixed emulsions or DOTAP-alone emulsions. The cytotoxicities of the DOTAP:DMPC mixed emulsions were 12.3%±2.3%, using 10 mL of the optimized DOTAP:DMPC mixed emulsions, a dose corresponding to 3.5 nmol lipid per 105 pfu Ad-GFP, significantly lower than the value, 42.6%±4.6%, obtained using the same dose of the DOTAP-alone emulsion (p<0.005) (Figure 3B). Taken together, an optimal balance between the cytotoxicity and the efficacy of viral gene transfer was obtained using the DOTAP:DMPC mixed emulsions prepared using a 3:3:6:0.06 mixture of DOTAP:DMPC:CHOL:PEG-PE and 17 μL/mL Lipiodol.
Physicochemical characteristics of the virus–emulsion complexes
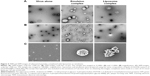
The viral gene transfer efficacies of the formulated DOTAP:DMPC mixed emulsions were further investigated by microscopy imaging to characterize the sizes and shapes of the virus–emulsion complexes. Images of the viral complexes prepared using 3:3 DOTAP:DMPC mixed liposomes that differed only in the absence of Lipiodol were obtained for comparison purposes.
TEM images revealed that the naked Ad formed a uniform spherical structure (100 nm in diameter), consistent with numerous previous studies, including ours.12,25 TEM analysis of the viral particles complexed with an emulsion in the ratio used for the gene transfer studies revealed a population of heterogeneous spherical vesicles (50-500 nm) in the images. Surprisingly, free viral particles and viral particles attached to emulsions were barely detectable in this sample, indicating that the viral particles were completely shielded by the emulsions (Figure 4A and B). By contrast, a TEM image of the virus–liposome complexes revealed that the viral particles were readily detected as aggregates or as free particles, a limited number of liposomes (the spherical structures 200–450 nm in diameter) were detected, and the liposomal surface-attached viral particles were rare.
Scanning electron microscopy (SEM) analysis was performed to compare the viral complexes with the liposomes or emulsions. Imaging viral complexes with liposomes or emulsions revealed the presence of larger complexes in the large-area images, and these complexes were further examined (Figure 4C). The SEM images of the viral particles were similar to the TEM images in terms of particle shape, uniformity, and diameter. SEM images of the virus–emulsion complexes revealed spherical complexes. Neither free viral particles nor those attached to the surfaces of the emulsions were significantly detected in the SEM images of the virus–emulsion complexes. By contrast, heterogeneous cluster structures containing liposomes and viral aggregates consisting of large spherical liposomes attached to several small spherical viral particles were found in the SEM images of virus–liposome complexes (Figure 4C).
AFM images shown in Figure 5 confirmed that the naked Ad particles formed spherical structures 80 nm in diameter. Images of the virus–emulsion complexes revealed spherical structures, as observed in the TEM and SEM images. A cross-sectional analysis revealed that the surfaces of the virus–emulsion complexes were asymmetrical and bumpy, suggesting that the structures were generated by the fusion of at least 2 different particles. AFM images of the liposome–virus complexes revealed a cluster structure composed of a large central particle (liposome) and smaller pendant particles (viral particles). The heights of the viral complexes in either the liposomes or emulsions obtained from a cross-sectional analysis were 130-160 nm, much smaller than the diameters estimated from the TEM and SEM images. The size differences may have been due to structural deformations of the lipid-based nanoparticles from spherical vesicles to flattened vesicles, as is frequently observed in AFM images due to the adsorption of nanoparticles onto the mica substrate prior to imaging.27,28
Dynamic light scattering (DLS) analysis was performed to compare the particle size distribution, the mean size and surface charge on the DOTAP:DMPC mixed liposomes or emulsions in suspension, before and after viral complexation (Figure 6). The mean size of the DOTAP:DMPC mixed emulsions was much larger than that of the DOTAP emulsions16 and approximately half of the mean size of the DOTAP:DMPC mixed liposomes (360 vs 825 nm, Figure 6A and B). The surface charges of the DOTAP:DMPC mixed liposomes or emulsions were highly positive to a similar extent (+65.5 vs +70.0 mV, Figure 6C). Incubation of the Ad with the liposomes produced much larger particles (825-1,148 nm), the surface charges of which were less positive (between +70.0 and +58.4 mV), suggesting that the negative surface charges on the viral particles were only partially shielded. By contrast, the incubation of Ads with emulsions produced only slightly larger particles (360-387 nm) with surface charges that were slightly more positive (between +65.5 and +66.4 mV). Considering the negative surface charges on the Ad (−19 mV),16 the surfaces of the viral particles appeared to be completely shielded by the DOTAP:DMPC emulsions.
The AFM, TEM, SEM, and DLS studies revealed that the viral particles were wrapped inside the DOTAP:DMPC mixed emulsion particles, whereas they were simply associated with the surfaces of the liposomal counterpart. As the viral particles attach to the DOTAP:DMPC mixed emulsion surfaces by interaction with DOTAP (major interaction), PC and CHOL (minor interaction) on the surface, DOTAP molecules present inside the emulsions may have pulled the viral particles into the core of emulsion via electrostatic attraction and bound to the viral particles tightly. The presence of Lipiodol in the inner oil core of emulsions may have oriented the positively charged DOTAP molecules to facilitate their interactions with the negatively charged viral capsid surfaces.
Gene transfer properties of cationic lipid:phosphatidylcholine mixed emulsions in various cancer cell lines and serum conditions
Serum proteins often bind to gene delivery vectors complexed with cationic lipids or polymers, which interferes with complex-mediated gene transfer.12,29 The serum compatibility of the emulsions was tested by measuring GFP expression in B16-F10 cells after mixing with virus-DOTAP:DMPC emulsion complexes in the presence of various serum concentrations. Media containing 10% serum are commonly used for cell culture, and 50% serum conditions closely mimic physiological conditions.30 Figure 7A shows that the GFP expression levels in B16-F10 cells treated with the virus-DOTAP:DMPC emulsion complexes were similar (80%−86%), regardless of the serum content. These results indicated that the serum did not interfere with the complex-mediated gene transfer, suggesting that the Ad-DOTAP:DMPC mixed emulsions were highly stable in the presence of serum.
The enhanced gene transfer effects obtained from cationic lipid-based nanoparticles may depend on the CAR expression level. Figure 7B indicates that pre-complexation of Ad particles with the DOTAP:DMPC emulsion increased transgene expression by factors of 41.3 and 28.9 in HCT116 and H460 cells, respectively, in the presence of 10% serum. Both of these cell lines are adenoviral infection-sensitive due to a high CAR surface expression (2.1% vs 86.7% in HCT 116 cells and 2.99% vs 86.3% in H460 cells).7,31 In EA.hy926 cells, which are resistant to adenoviral infection due to the absence of CAR,16,32,33 pre-complexation of Ad particles with DOTAP:DMPC emulsion increased transgene expression by a factor of 34.2 (2.6% vs 88.8%). These data demonstrated the abilities of the DOTAP:PC mixed emulsions to enhance adenoviral gene transfer in a wide range of cell types. Delivering viral particles in cationic lipid:phosphatidylcholine mixed emulsions may reduce repulsion among negatively charged Ad particles and negatively charged cell membranes, thereby increasing the attachment and entry of viral particles into the cells, regardless of the CAR expression level.
Conclusion
In the present study, cationic lipid:phosphatidylcholine mixed nanoparticles were successfully formulated as a highly effective vehicle for enhancing adenoviral gene transfer into cells. The formula was optimized by 1) substituting a portion of the cationic lipid DOTAP with a zwitterionic lipid DMPC, and 2) optimizing the inner oil Lipiodol content. The formulated DOTAP:DMPC mixed emulsions exhibited a strong capacity for enhancing adenoviral gene transfer in a broad range of cells, even in the presence of high serum levels, while producing less cytotoxicity compared with the DOTAP emulsions. Importantly, we explored the structural basis for the effectiveness of the optimized DOTAP:DMPC mixed emulsions compared with liposomal counterpart: the DOTAP:DMPC mixed emulsions, if they contain DOTAP at the minimum required content, or more, spontaneously surrounded the Ad particles and encapsulated the viral particles within the emulsions. These studies demonstrated that optimized DOTAP:DMPC mixed emulsions are potentially useful for adenoviral gene delivery due to less cytotoxicity and the unique ability to encapsulate the viral particle, highlighting the importance of nanoparticle formulation.
Acknowledgments
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (No 2015R1A2A2A01005783) and a grant (16173MFDS542) from Ministry of Food and Drug Safety in 2017.
Disclosure
The authors report no conflicts of interest in this work.
References
Kang E, Yun CO. Current advances in adenovirus nanocomplexes: more specificity and less immunogenicity. BMB Rep. 2010;43(12):781–788. | ||
Yoon AR, Hong J, Kim SW, Yun CO. Redirecting adenovirus tropism by genetic, chemical, and mechanical modification of the adenovirus surface for cancer gene therapy. Expert Opin Drug Deliv. 2016;13(6):843–858. | ||
Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2003;5(6):451–462. | ||
Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012 – an update. J Gene Med. 2013;15(2):65–77. | ||
Krasnykh VN, Douglas JT, van Beusechem VW. Genetic targeting of adenoviral vectors. Mol Ther. 2000;1(5 Pt 1):391–405. | ||
Kawabata K, Sakurai F, Koizumi N, Hayakawa T, Mizuguchi H. Adenovirus vector-mediated gene transfer into stem cells. Mol Pharm. 2006;3(2):95–103. | ||
Lee EM, Hong SH, Lee YJ, et al. Liposome-complexed adenoviral gene transfer in cancer cells expressing various levels of coxsackievirus and adenovirus receptor. J Cancer Res Clin Oncol. 2004;130(3):169–177. | ||
Han SY, Lee YJ, Jung HI, et al. Gene transfer using liposome-complexed adenovirus seems to overcome limitations due to coxsackievirus and adenovirus receptor-deficiency of cancer cells, both in vitro and in vivo. Exp Mol Med. 2008;40(4):427–434. | ||
Zhao C, Wu N, Deng F, et al. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS One. 2014;9(3):e92908. | ||
Kim SY, Lee MK, Lim SY. Current advances in developing cationic lipid-based nanoparticles as a vehicle for improving adenoviral gene delivery. J Pharm Investig. 2016;46(4):393–402. | ||
Park JY, Kim MG, Shim GY, Oh KY. Lipid-based antigene delivery systems. J Pharm Investig. 2016;46(4):295–304. | ||
Kim SY, Lee SJ, Han HK, Lim SJ. Aminoclay as a highly effective cationic vehicle for enhancing adenovirus-mediated gene transfer through nanobiohybrid complex formation. Acta Biomater. 2017;49:521–530. | ||
Chan CL, Ewert KK, Majzoub RN, et al. Optimizing cationic and neutral lipids for efficient gene delivery at high serum content. J Gene Med. 2014;16(3–4):84–96. | ||
Zhang S, Xu Y, Wang B, Qiao W, Liu D, Li Z. Cationic compounds used in lipoplexes and polyplexes for gene delivery. J Control Release. 2004;100(2):165–180. | ||
Worgall S, Worgall TS, Kostarelos K, et al. Free cholesterol enhances adenoviral vector gene transfer and expression in CAR-deficient cells. Mol Ther. 2000;1(1):39–48. | ||
Kim SY, Lee SJ, Lim SJ. Formulation and in vitro and in vivo evaluation of a cationic emulsion as a vehicle for improving adenoviral gene transfer. Int J Pharm. 2014;475(1–2):49–59. | ||
Yotnda P, Chen DH, Chiu W, et al. Bilamellar cationic liposomes protect adenovectors from preexisting humoral immune responses. Mol Ther. 2002;5(3):233–241. | ||
Qiu C, De Young MB, Finn A, Dichek DA. Cationic liposomes enhance adenovirus entry via a pathway independent of the fiber receptor and alpha(v)-integrins. Hum Gene Ther. 1998;9(4):507–520. | ||
Bashyal S, Noh GB, Keum TW, Choi YW, Lee SK. Cell penetrating peptides as an innovative approach for drug delivery; then, present and the future. J Pharm Investig. 2016;46:205–220. | ||
Lee YS, Kim SW. Bioreducible polymers for therapeutic gene delivery. J Ontrol Release. 2014;190:424–439. | ||
Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114(1):100–109. | ||
Soenen SJ, Brisson AR, De Cuyper M. Addressing the problem of cationic lipid-mediated toxicity: the magnetoliposome model. Biomaterials. 2009;30(22):3691–3701. | ||
Wei X, Shao B, He Z, et al. Cationic nanocarriers induce cell necrosis through impairment of Na(+)/K(+)-ATPase and cause subsequent inflammatory response. Cell Res. 2015;25(2):237–253. | ||
Balakireva L, Schoehn G, Thouvenin E, Chroboczek J. Binding of adenovirus capsid to dipalmitoyl phosphatidylcholine provides a novel pathway for virus entry. J Virol. 2003;77(8):4858–4866. | ||
Singh R, Al-Jamal KT, Lacerda L, Kostarelos K. Nanoengineering artificial lipid envelopes around adenovirus by self-assembly. ACS Nano. 2008;2(5):1040–1050. | ||
Lechanteur A, Furst T, Evrard B, Delvenne P, Hubert P, Piel G. PEGylation of lipoplexes: The right balance between cytotoxicity and siRNA effectiveness. Eur J Pharm Sci. 2016;93:493–503. | ||
Nakano K, Tozuka Y, Yamamoto H, Kawashima Y, Takeuchi H. A novel method for measuring rigidity of submicron-size liposomes with atomic force microscopy. Int J Pharm. 2008;355(1–2):203–209. | ||
Et-Thakafy O, Delorme N, Gaillard C, et al. Mechanical properties of membranes composed of gel-phase or fluid-phase phospholipids probed on liposomes by atomic force spectroscopy. Langmuir. 2017;33(21):5117–5126. | ||
Uchida E, Mizuguchi H, Ishii-Watabe A, Hayakawa T. Comparison of the efficiency and safety of non-viral vector-mediated gene transfer into a wide range of human cells. Biol Pharm Bull. 2002;25(7):891–897. | ||
Zhang Y, Arrington L, Boardman D, et al. The development of an in vitro assay to screen lipid based nanoparticles for siRNA delivery. J Control Release. 2014;174:7–14. | ||
Bagheri N, Shiina M, Lauffenburger DA, Korn WM. A dynamical systems model for combinatorial cancer therapy enhances oncolytic adenovirus efficacy by MEK-inhibition. PLoS Comput Biol. 2011;7(2):e1001085. | ||
Nettelbeck DM, Miller DW, Jerome V, et al. Targeting of adenovirus to endothelial cells by a bispecific single-chain diabody directed against the adenovirus fiber knob domain and human endoglin (CD105). Mol Ther. 2001;3(6):882–891. | ||
Preuss MA, Glasgow JN, Everts M, Stoff-Khalili MA, Wu H, Curiel DT. Enhanced gene delivery to human primary endothelial cells using tropism-modified adenovirus vectors. Open Gene Ther J. 2008;1:7–11. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.