Back to Journals » OncoTargets and Therapy » Volume 12
Oncogenic Role and Prognostic Value of MicroRNA-937-3p in Patients with Breast Cancer
Authors Li D, Zhong J, Zhang G, Lin L, Liu Z
Received 2 September 2019
Accepted for publication 16 November 2019
Published 13 December 2019 Volume 2019:12 Pages 11045—11056
DOI https://doi.org/10.2147/OTT.S229510
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Deyu Li,1,2 Jiangming Zhong,1,2 Guifeng Zhang,1,2 Li Lin,1,2 Zhenhua Liu1,2
1Department of Medical Oncology, Provincial Clinical College, Fujian Medical University, Fuzhou 350001, People’s Republic of China; 2Department of Medical Oncology, Fujian Provincial Hospital, Fuzhou 350001, People’s Republic of China
Correspondence: Zhenhua Liu
Department of Medical Oncology, Provincial Clinical College, Fujian Medical University, No. 134 Dongjie Street, Fuzhou 350001, Fujian Province, People’s Republic of China
Tel +86 591-88217461
Email [email protected]
Purpose: Breast cancer is the most common female tumor in the world. MicroRNA has been reported to play an important role in the progression of breast cancer. The purpose of this study was to explore the role of miR-937-3p in breast cancer.
Patients and methods: Expression of miR-937-3p in breast cancer tissues and serums was detected from The Cancer Genome Atlas (TCGA), the Gene Expression Omnibus (GEO) and patients’ samples. Kaplan–Meier plotter identified the association between miR-937-3p and prognosis.
Results: The analysis of TCGA, GEO and qRT-PCR suggested that the level of miR-937-3p was increased in breast cancer tissues and serum compared with adjacent normal breast tissues and healthy persons, respectively. The decreased expression of miR-937-3p inhibited breast cancer proliferation, migration and invasion. CCRL2 was the target of miR-937-3p. In contrast to miR-937-3p, the level of CCRL2 was decreased in breast cancer tissues. Luciferase reporter assay revealed that miR-937-3p directly bound to the 3ʹ-UTR of CCRL2. Double knockdown of CCRL2 and miR-937-3p promoted breast cancer cell proliferation, migration and invasion, suggesting that miR-937-3p promoted breast cancer cell proliferation, migration and invasion by targeting CCRL2. The Kaplan–Meier survival analysis suggested that breast cancer patients with high level of miR-937-3p or low level of CCRL2 had a reduced overall survival (OS).
Conclusion: miR-937-3p plays an important role in the diagnosis and prognosis of breast cancer. Inhibition of miR-937-3p expression may be a novel targeted therapy for breast cancer.
Keywords: miR-937-3p, breast cancer, proliferation, migration, invasion, prognosis
Introduction
The incidence of cancer in the world is increasing year by year, and cancer has replaced diseases such as heart disease as the main cause of human death.1 Breast cancer is one of the most commonly diagnosed malignant tumors in women. In recent years, the incidence of breast cancer has gradually increased, and the age of onset has gradually become younger, which poses a great threat to women’s health.2 Tumor formation is a complex dynamic process involving multiple biological factors.
MicroRNAs are a class of non-coding RNAs of 20–22 nucleotides, and able to couple with its complementary mRNA, leading to degradation of the coupled mRNA.3–7 Lee et al first discovered the first gene lin-4 that regulates late embryo development in Caenorhabditis elegans, which opened the miRNA research prelude.8 Related studies have shown that miRNAs that regulate gene expression are involved in almost every cellular process, including cholesterol metabolism, embryo implantation, insulin synthesis, and hematopoiesis.9–12 miRNAs play an important role in the regulation of biological processes such as cell proliferation, cell pluripotency, tissue differentiation, and apoptosis.13,14 Abnormal expression of miRNAs can cause a variety of diseases. It has become an indisputable fact that miRNAs are abnormally expressed in most malignant tumors in humans. Specific tumor types have different miRNA expression, and miRNA expression profiling can be used to identify the major tissue origin of the tumor.15 Previous study found that at least half of miRNAs were first expressed in gene expression profiles associated with cancer.16 In the process of tumor formation, the dysregulation of miRNAs may lead to increased translation of oncoproteins and/or tumor suppressor proteins.
Abnormal expression of miRNA was found in several different tumor tissues compared with corresponding normal tissues. A class of miRNAs is lowly expressed in normal tissues and highly expressed in tumor tissues, and is called a “cancerous miRNA”. For example, He et al found that miRNA-146b, miRNA-221 and miRNA-222 are abnormally highly expressed in thyroid malignancies, and the target gene of these miRNAs is c-KIT gene that play an important role in cell growth and differentiation.17 In contrast, another class of miRNAs that are highly expressed in normal tissues and low in tumor tissues are called “cancer suppressor miRNAs”. A typical example of a miRNA that plays a role in tumor suppression in the development of tumors is the let-7 gene.18 The specificity of miRNAs in tumors can help us distinguish between cancer cells and normal cells, and it can play a significant role in predicting the efficacy of specific drugs or the prognosis of patients. Dysregulation of various microRNAs has been demonstrated in breast cancer, including miR-10b, miR-373, miR-520, miR-335, miRNA-126 and miR-206, which associated with the development and progression of breast cancer.19–23 MiR-937 has been reported to play an important role in the development of various types of diseases, including lung cancer, gastric cancer, and Alzheimer’s disease.24–26 However, it remains unclear whether miR-937-3p is involved in the development and progression of breast cancer.
Previous studies reported that miR-937 has various functions in different types of diseases, is one of the miRNAs related to diseases. miR-937 acts as a suppressive factor in gastric cancer by downregulating Forkhead box protein L2 (FOXL2).25 However, it is an oncogene in lung cancer cell proliferation by targeting inositol polyphosphate-4-phosphatase type II (INPP4B).24 However, the expression of miR-937-3p in breast cancer and its effect on breast cancer cell proliferation and metastasis, as well as the relationship between miR-937-3p and clinical parameters, has not been reported.
This study investigated the correlations between miR-937-3p expression, cell function and clinical characteristics through data collected from Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) database and RT-PCR to determine the clinical role of miR-937-3p in breast cancer, and cell function analyses to determine the role of miR-937-3p in breast cancer cell.
Materials and Methods
Clinical Tissue Specimens
A total of 20 pairs of fresh breast cancer tissue and adjacent normal tissue were collected from cases who underwent curative resection at the Fujian Provincial Hospital. Fresh clinical tissue specimens were immediately snap-frozen in liquid nitrogen for RNA extraction. All breast cancer clinical tissue specimens were histologically diagnosed and confirmed by clinical pathologists. Twenty serum samples of healthy control and 20 serum samples of breast cancer patients were collected from Fujian Provincial Hospital. The Ethics Committee of the Fujian Provincial Hospital has approved this experimental scheme. The written informed consent was obtained from all breast cancer cases.
Cell Culture
The breast cancer cell lines MCF-7, MDA-MB-231, HCC1954 and the non-transformed mammary epithelial MCF-10A cells were obtained from the ATCC (Manassas, VA, USA). MCF-7 cells were cultured in DMEM (Invitrogen, Carlsbad, CA) supplemented with 1% PS (penicillin and streptomycin) and 10% fetal bovine serum (FBS). MDA-MB-231 cells and HCC1954 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 1% PS and 10% FBS. MCF-10A cells were cultured in Dulbecco’s modified Eagle's medium supplemented with 1% PS and 10% FBS. Cells were grown at 37°C in the presence of 5% CO2.
Oligonucleotides and Transfection
miR-937-3p-mimic, miR-937-3p-Mut, miR-937-3p-inhibitor, CCRL2-siRNA and their negative control were purchased from Genepharma (Genepharma, Shang Hai, China), and transfected into MCF-7 cells using lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s guidelines.
Western Blot
The cells were lysed in lysis buffer for 30 mins and centrifuged at 4°C, 13,000 rpm for 10 mins to remove insoluble material. The soluble protein concentration was determined by Bradford assay. Protein samples were separated by SDS-PAGE and transferred to nitrocellulose blotting membranes (Bio-Rad, CA, USA). The membranes were treated with block buffer at room temperature for 1 hr. The membranes were then incubated with primary antibodies overnight at 4°C, then washing in PBS/Tween-20, followed by incubating with secondary antibodies in blocking buffer at room temperature for 2 hrs. Finally, washing in PBS/Tween-20 again. The signals were detected and measured using LICOR Odyssey system (LI-COR, NE, USA). Anti-GAPDH antibody (60004-1-Ig) and anti-CCRL2 antibody (66611-1-Ig) were purchased from Proteintech (Wuhan, China).
RNA Extraction and Quantitative Polymerase Chain Reaction
Total RNA was extracted from cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. TaqMan MicroRNA Reverse Transcription kit (Takara, Otsu, Japan) was used to synthesize cDNA from total RNA at 37°C for 30 mins. qPCR was performed on an Applied Biosystems StepOne Plus Real-Time PCR System (Takara, Otsu, Japan). All experiments were performed in at least triplicate. Relative quantification of miR-973-3p expression and CCRL2 in cell lines was calculated with the 2−∆∆Ct method. Relative quantification of miR-973-3p expression in patients' tissue and serum was calculated with the 2−∆Ct method. The relative levels of miR-973-3p in patients' tissue and serum were calculated using the comparative 2−∆Ct method relative to U6 and cel-miR-39.
Luciferase Reporter Assay
The fragment of the 3ʹ-UTR of CCRL2 containing the binding site for miR-937-3p was cloned into the pGL3 vector and cotransfected with the miR-937-3p mimic or miR-937-3p Mut mimic into the cells using Lipofectamine 2000. Luciferase reporter assays were performed after 48 hrs of posttransfection according to the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was normalized to Renilla luciferase activity.
Cell Proliferation Assay
Cells were seeded in 96-well culture plates at a density of 3000–8000 cells/well. On the following day, the cells were exposed to various concentrations of compounds and were cultured for another 72 h. Cell proliferation was then determined by sulforhodamine B (SRB) or with the Cell Counting Kit-8 (CCK8) assay. The absorbance was determined at 515 nm or 450 nm.
Cell Migration and Cell Invasion Assay
Cell migration assay was detected by scratch assay. Scratches were made by using a 100 μL pipette tip at 80% cells confluence post-transfection. The wound closure was observed by exhaustively washed with PBS and the migration rate was calculated according to the ratio of migrated cell surface area to total surface area.
Cell invasion assays were performed with Transwell chamber. The cells (5 × 104) were seeded in the interior of the insert chamber (Corning) with the Matrigel-coated membrane. After fixation and staining with 0.1% crystal violet, five random fields under microscope for each group were selected and counted.
Bioinformatics Analysis
The potential target genes of miR-937-3p were predicted using miRDB (http://www.mirdb.org/) and TargetScan (http://www.Targetscan.org/). The predicted target genes supported by both methods were selected for further analysis.
Statistical Analysis
In the research, the Student’s t-test was used to calculate the miRNA or mRNA expression level by using GraphPad-prism5. Log-rank test was used to calculate the survival analysis by using IBM SPSS version 19.0. Multivariate survival analysis was performed by using stepwise Cox proportional hazards regression model. All p value of < 0.05 was considered statistically significant.
Results
The Level of miR-937-3p Was Increased in Breast Cancer Tissues
In order to explore the expression of miR-937-3p in breast cancer tissues, TCGA and GEO database was employed. Based on TCGA, GSE42072 and GSE58606 dataset, we found that the level of miR-937-3p was increased in breast cancer tissues compared with adjacent normal breast tissues (Figure 1A–C). And then, 20 samples of breast cancer tissues and 20 samples of adjacent normal tissues were applied and further confirm the conclusion that the level of miR-937-3p was increased in breast cancer tissues (Figure 1D).
There was a significant difference in miR-937-3p expression between breast cancer and normal breast tissues. Receiver operating characteristic (ROC) curve analysis showed good diagnostic values for breast cancer, including area under the curve (AUC) values. Data from the TCGA dataset showed that miR-937-3p expression was a significant diagnostic factor for breast cancer (AUC=0.758; 95% CI, 0.720–0.795) (Figure 1E). For the remaining two datasets studied, the AUC was 0.959 in the GSE42072 dataset (Figure 1F), 0.846 in the GSE58606 dataset (Figure 1G). For the 20 pairs of breast cancer and normal tissues, the AUC was 0.837 (Figure 1H). Based on the above data, miR-937-3p expression might be used as a new biomarker to distinguish between breast cancer tissue and matched adjacent normal breast tissue.
The Level of Serum miR-937-3p Was Increased in Breast Cancer Patients Compared with Healthy Person
According to the data from the GSE73002, we found that the level of serum miR-937-3p was significantly increased in breast cancer patient compared with healthy person (Figure 2A). Meanwhile, serum miR-937-3p levels in 20 samples of breast cancer patients and 20 samples of healthy persons were detected by RT-qPCR. The results showed that serum miR-937-3p was significantly up-regulated in breast cancer patients compared to healthy controls (Figure 2B). ROC curve was used to analyze the ability of serum miR-937-3p to discriminate between breast cancer patients or healthy control. For the GSE73002 dataset. The AUC of serum miR-937-3p was 0.904 (Figure 2C). For the 20 samples of breast cancer patients and 20 samples of healthy persons, the AUC was 0.914 (Figure 2D), suggesting serum miR-937-3p can help distinguish breast cancer patients. Based on the above data, serum miR-937-3p expression might be used as a new biomarker to distinguish between breast cancer and healthy persons.
Down-Regulation of miR-937-3p Inhibited the Proliferation, Migration and Invasion of Breast Cancer Cell
To investigate the role of miR-937-3p in breast cancer, RT-qPCR was used to quantify the expression of miR-937-3p in different cell lines including breast cancer cell lines MDA-MB-231, HCC1954, MCF-7 and normal immortalized MCF-10A breast cell line. RT-qPCR analysis demonstrated that the level of miR-937-3p was significantly increased in breast cancer cell lines compared with breast cell line MCF-10A (Figure 3A). In order to investigate the functions of miR-937-3p in breast cancer, MCF-7 cells were transfected with miR-937-3p inhibitor or miR-937-3p-NC. The efficiency of the transfection was determined by RT-qPCR. As demonstrated in Figure 3B, the level of miR-937-3p was significantly decreased in miR-937-3p inhibitor transfected MCF-7 cells compared with cells transfected with miR-937-3p-NC.
Cell proliferation assay was performed to investigate the effect of miR-937-3p on the proliferation of MCF-7 cells. As demonstrated in Figure 3C and D, the decreased level of miR-937-3p significantly inhibits cell proliferation in MCF-7 cells. And then, we used wound healing assay (Figure 3E and F) and Transwell chamber (Figure 3G and H) to detect the effect of miR-937-3p on MCF-7 cells migration and invasion abilities. These results indicated that miR-937-3p-inhibitor transfection cells had a significant inhibition of cell migration and invasion in MCF-7 cells.
miR-937-3p Negatively Regulates the Expression of CCRL2 by Directly Binding to Its 3′-UTR
In order to find a potential target gene of miR-937-3p that may contribute to its functions in breast cancer cells, bioinformatic analysis was performed. As presented in Figure 4A–C, there was complementarity between miR-937-3p and the 3′UTR region of CCRL2. Based on TCGA dataset and GSE70905 dataset, we found that the level of CCRL2 was decreased in breast cancer tissues compared with adjacent normal breast tissues (Figure 4D and E). Using 20 freshly collected clinical breast cancer samples, we found that miR-937-3p expression correlated inversely with the levels of CCRL2 (Figure 4F). The direct regulation of CCRL2 by miR-937-3p in the breast cancer cells was confirmed by luciferase reporter assay. The luciferase reporter assay demonstrated markedly decreased activity with miR-937-3p-mimics and CCRL2-3′UTR compared with the miR-937-3p-Mut and CCRL2-3′UTR (Figure 4G).
In addition, Western blot analysis was performed to confirm the regulatory effect of miR-937-3p on CCRL2 expression level in the breast cancer cells. As demonstrated in Figure 4H and I, miR-937-3p mimics decreased the CCRL2 expression level in the breast cancer cells at the protein levels. Taken together, these results indicated that miR-937-3p negatively regulated CCRL2 expression by directly binding to its 3′UTR region in the breast cancer cells.
Inhibition of CCRL2 Is Important for miR-937-3p Induced Cell Proliferation, Migration and Invasion in Breast Cancer cell
The effect of CCRL2 reduction on breast cancer progression was examined by repressing endogenous CCRL2 expression using an CCRL2-specific siRNA (Figure 5A and B). As showed in Figure 5C–H, silencing CCRL2 in miR-937-3p-inhibitor transfected breast cancer cells increased the ability of cell proliferation, migration and invasion of breast cancer cells. These results suggested that further silencing CCRL2 expression in miR-937-3p-inhibitor cells could reverse the inhibitory effect of the miR-937-3p on breast cancer cells’ proliferation, migration and invasion. These data confirmed that miR-937-3p promoted breast cancer cells proliferation, migration and invasion by repressing endogenous CCRL2 expression and that CCRL2 plays an important role in miR-937-3p-mediated breast cancer cell proliferation, migration and invasion.
The Prognostic Value of miR-937-3p and CCRL2 mRNA Expression in Breast Cancer
In the TCGA BRCA cohort, we observed that patients with high expression level of miR-937-3p were at significantly increased risk of death (Figure 6A), advanced stage and patients with age >60 also have a high risk of death (Table 1). The website of the Kaplan–Meier Plotter was used to evaluate the prognostic value of CCRL2 mRNA in breast cancer. Kaplan–Meier survival analysis based on GSE16446 showed that low level of CCRL2 correlated with an unfavorable clinical outcome compared with patients with breast cancer and high level of CCRL2 (Figure 6B). Further analysis showed that the breast cancer patients with high level of miR-937-3p and low level of CCRL2 had a reduced OS compared with the breast cancer patients with low level of miR-937-3p and high level of CCRL2 based on TCGA BRCA cohort (Figure 6C). Therefore, the decreased levels of miR-937 might be a protective factor for patients with breast cancer.
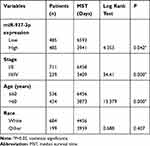 |
Table 1 Univariate Analysis of the Correlation Between Clinicopathological Parameters and Survival of BRCA Patients in TCGA Cohort |
Discussion
In recent years, the incidence of breast cancer has continued to increase in most Asian countries. Early diagnosis and comprehensive treatment have a significant effect on the survival time and quality of life in breast cancer patients. At present, the treatment of breast cancer is mainly based on surgery, combined with radiotherapy, chemotherapy, endocrine therapy and molecular targeted therapy. As the disease progresses or resolves, there are still significant and unpredictable results in clinical performance between different patients.
In the pathological and physiological processes of different diseases, miRNA plays an important role in gene management by regulating the expression of thousands of target genes through post-transcriptional levels. Most miRNAs are located in introns and function together with their host genes for transcription. Deregulated miRNAs not only affect cancer development, invasion and metastasis, but are also associated with the development of resistance to a variety of anticancer drugs. Due to the specific expression pattern of miRNAs in tissues or diseases and the ability to regulate gene expression, miRNAs have been identified as biomarkers for the diagnosis and prognosis, and have also been accepted as a novel therapeutic approach.
MiR-937 has been confirmed to be involved in cell proliferation and metastasis. Yu et al found that the level of miR-937 was decreased in gastric cancer cells and tissues. Further research found that miR-937 inhibits the proliferation and metastasis of gastric cancer cells by targeting FOXL2 via inactivation of PI3K/AKT signaling pathway. However, the level of miR-937 was increased in lung cancer cells and tissues, and miR-937 contributed to cell proliferation of lung cancer by inhibiting INPP4B.
Our research showed that the level of miR-937-3p was increased in breast cancer tissue, blood and cells. At the same time, miR-937-3p played as an oncogenic role in regulating breast cancer cells proliferation, migration and invasion. We found that miR-937-3p directly targeted 3ʹ-UTR of CCRL2, and negatively regulated CCRL2 expression in MCF-7 cell. For exploring the role of CCRL2 in miR-937-3p induced cell proliferation, migration and invasion in breast cancer cells, we conducted the rescued-function study and observed that double knockdown of CCRL2 and miR-937-3p promoted breast cancer cell proliferation, migration and invasion. Therefore, miR-937-3p promoted cell proliferation, migration and invasion through regulating the CCRL2 expression in breast cancer cells.
The clinical and prognostic significance of miR-937-3p was seldom reported in human cancers. In the research, we found that the level of miR-937-3p expression was increased in the serum of breast cancer patients compared with healthy persons. Meanwhile, we found that breast cancer patients with high level of miR-937-3p were at significantly higher risk of death. Therefore, miR-937-3p plays an important role in the diagnosis and prognosis of breast cancer.
Conclusions
In this experiment, RT-PCR and cell function experiments were used to confirm that mir-937-3p is highly expressed in breast cancer tissues, and it is involved in the growth of breast cancer tumors as a tumor-promoting gene. Inhibition of miR-937-3p expression may become a novel targeted therapy for breast cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pierce BA. CA: a cancer journal for clinicians. Biomed Market Newsl. 2012.
2. Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: current perspectives. Breast Cancer. 2014;2014:1.
3. Zhang S, Liu L, Lv Z, Li Q, Gong W, Wu H. MicroRNA-342-3p Inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1). Oncol Res. 2017;25(9):1505. doi:10.3727/096504017X14886485417426
4. Zhang X, Gao F, Zhou L, Wang H, Shi G, Tan X. UCA1 regulates the growth and metastasis of pancreatic cancer by sponging MiR-135a. Oncol Res. 2017;25(9):1529–1541. doi:10.3727/096504017X14888987683152
5. Jiang Z, Yao L, Ma H, et al. miRNA-214 inhibits cellular proliferation and migration in glioma cells targeting caspase-1 involved in pyroptosis. Oncol Res. 2016;25(6):1009. doi:10.3727/096504016X14813859905646
6. Li X, Li Y, Lu H. MiR-1193 suppresses proliferation and invasion of human breast cancer cells through directly targeting IGF2BP2. Oncol Res. 2017;25(4):579–585. doi:10.3727/97818823455816X14760504645779
7. Jiang Z, Zhang Y, Cao R, et al. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017;25(7):1081–1087. doi:10.3727/096504016X14831120463349
8. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843. doi:10.1016/0092-8674(93)90529-Y
9. Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21(12):699–706. doi:10.1016/j.tem.2010.08.008
10. Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138(9):1653. doi:10.1242/dev.056234
11. Melkman-Zehavi T, Oren R, Kredo-Russo S, et al. miRNAs control insulin content in pancreatic β‐cells via downregulation of transcriptional repressors. EMBO J. 2011;30(5):835–845. doi:10.1038/emboj.2010.361
12. O’Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood. 2011;118(11):2960–2969. doi:10.1182/blood-2011-03-291971
13. Zhou Z, Zhou L, Jiang F, et al. Downregulation of miR-222 induces apoptosis and cellular migration in adenoid cystic carcinoma cells. Oncol Res. 2017;25(2):207–214. doi:10.3727/096504016X14732772150460
14. Wang J, Sun G. FOXO1-MALAT1-miR-26a-5p feedback loop mediates proliferation and migration in osteosarcoma cells. Oncol Res. 2017;25(9):1517–1527. doi:10.3727/096504017X14859934460780
15. Ferracin M, Pedriali M, Veronese A, et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43–53. doi:10.1002/path.2915
16. Sevignani C, Calin GA, Nnadi SC, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci U S A. 2007;104(19):8017–8022. doi:10.1073/pnas.0702177104
17. He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–19080. doi:10.1073/pnas.0509603102
18. Pu XX, Huang GL, Guo HQ, et al. Circulating miR‐221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–1680. doi:10.1111/j.1440-1746.2010.06417.x
19. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi:10.1038/nature06174
20. Hafez MM, Hassan ZK, Zekri AR, et al. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pac J Cancer Prev. 2012;13(2):591–598. doi:10.7314/APJCP.2012.13.2.591
21. Sahin Ö, Schneeweiss A, Shavinskaya A, et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. 2012;31(37):4150. doi:10.1038/onc.2011.571
22. Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi:10.1038/ncb1681
23. Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol. 2009;2(4):361–369.
24. Zhang L, Zeng D, Chen Y, et al. miR-937 contributes to the lung cancer cell proliferation by targeting INPP4B. Life Sci. 2016;155:110–115. doi:10.1016/j.lfs.2016.05.014
25. Yu L, Chen J, Liu Y, Zhang Z, Duan S. MicroRNA-937 inhibits cell proliferation and metastasis in gastric cancer cells by downregulating FOXL2. Cancer Biomarkers. 2017;21(1):1–12. doi:10.3233/CBM-170310
26. Liu Z, Wang C, Wang X, Xu S. Therapeutic effects of transplantation of as-miR-937-expressing mesenchymal stem cells in murine model of Alzheimer’s disease. Cell Physiol Biochem. 2015;37(1):321–330. doi:10.1159/000430356
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






